Abstract
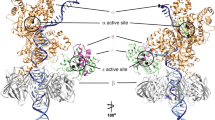
Almost all DNA polymerases show a strong preference for incorporating the nucleotide that forms the correct Watson–Crick base pair with the template base. In addition, the catalytic efficiencies with which any given polymerase forms the four possible correct base pairs are roughly the same. Human DNA polymerase-ι (hPolι), a member of the Y family of DNA polymerases, is an exception to these rules. hPolι incorporates the correct nucleotide opposite a template adenine with a several hundred to several thousand fold greater efficiency than it incorporates the correct nucleotide opposite a template thymine, whereas its efficiency for correct nucleotide incorporation opposite a template guanine or cytosine is intermediate between these two extremes1,2,3,4,5. Here we present the crystal structure of hPolι bound to a template primer and an incoming nucleotide. The structure reveals a polymerase that is ‘specialized’ for Hoogsteen base-pairing, whereby the templating base is driven to the syn conformation. Hoogsteen base-pairing offers a basis for the varied efficiencies and fidelities of hPolι opposite different template bases, and it provides an elegant mechanism for promoting replication through minor-groove purine adducts that interfere with replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S. & Prakash, L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406, 1015–1019 (2000)
Tissier, A., McDonald, J. P., Frank, E. G. & Woodgate, R. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 14, 1642–1650 (2000)
Zhang, Y., Yuan, F., Wu, X. & Wang, Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol. Cell. Biol. 20, 7009–7108 (2000)
Haracska, L. et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA 98, 14256–14261 (2001)
Washington, M. T., Johnson, R. E., Prakash, L. & Prakash, S. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 24, 936–943 (2004)
Prakash, S. & Prakash, L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16, 1872–1883 (2002)
Johnson, R. E., Prakash, S. & Prakash, L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase Polη. Science 283, 1001–1004 (1999)
Washington, M. T., Johnson, R. E., Prakash, S. & Prakash, L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl Acad. Sci. USA 97, 3094–3099 (2000)
Johnson, R. E., Washington, M. T., Prakash, S. & Prakash, L. Fidelity of human DNA polymerase η. J. Biol. Chem. 275, 7447–7450 (2000)
Washington, M. T., Prakash, L. & Prakash, S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Natl Acad. Sci. USA 100, 12093–12098 (2003)
Johnson, R. E., Kondratick, C. M., Prakash, S. & Prakash, L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285, 263–265 (1999)
Masutani, C. et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 (1999)
Trincao, J. et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8, 417–426 (2001)
Ling, H., Boudsocq, F., Woodgate, R. & Yang, W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 (2001)
Zhou, B.-L., Pata, J. D. & Steitz, T. A. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8, 427–437 (2001)
Silvian, L. F., Toth, E. A., Pham, P., Goodman, M. F. & Ellenberger, T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol. 8, 984–989 (2001)
Steitz, T. A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274, 17395–17398 (1999)
Faili, A. et al. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature 419, 944–947 (2002)
Leontis, N. B., Stombaugh, J. & Westhof, E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30, 3497–3531 (2002)
Johnson, R. E., Trincao, J., Aggarwal, A. K., Prakash, S. & Prakash, L. Deoxynucleotide triphosphate binding mode conserved in Y family DNA polymerases. Mol. Cell. Biol. 23, 3008–3012 (2003)
DeLucia, A. M., Grindley, N. D. & Joyce, C. M. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discriminating against ribonucleotides. Nucleic Acids Res. 31, 4129–4137 (2003)
Doublie, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 (1998)
Li, Y., Korolev, S. & Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525 (1998)
Washington, M. T. et al. Efficient and error-free replication past a minor groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 24, 5687–5693 (2004)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Acknowledgements
We thank staff at the APS for facilitating data collection; and T. Edwards and C. Escalante for help with structure determination. This work was supported by grants from the NIH (to A.K.A and L.P.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Table 1
Table listing data collection, phasing, and refinement statistics. (DOC 38 kb)
Supplementary Figure 1
A figure comparing Pol iota with Dpo4. (JPG 53 kb)
Supplementary Figure 1 Legend
A legend for Supplementary Figure 1. (DOC 19 kb)
Rights and permissions
About this article
Cite this article
Nair, D., Johnson, R., Prakash, S. et al. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 (2004). https://doi.org/10.1038/nature02692
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02692
This article is cited by
-
In crystallo observation of three metal ion promoted DNA polymerase misincorporation
Nature Communications (2022)
-
Structure of human telomerase holoenzyme with bound telomeric DNA
Nature (2021)
-
Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide
Nature Communications (2021)
-
Quaternary structural diversity in eukaryotic DNA polymerases: monomeric to multimeric form
Current Genetics (2020)
-
Structure and function relationships in mammalian DNA polymerases
Cellular and Molecular Life Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.