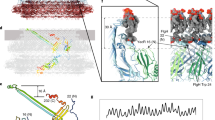
Abstract
The bacterial flagellar filament is a helical propeller constructed from 11 protofilaments of a single protein, flagellin. The filament switches between left- and right-handed supercoiled forms when bacteria switch their swimming mode between running and tumbling. Supercoiling is produced by two different packing interactions of flagellin called L and R. In switching from L to R, the intersubunit distance (∼52 Å) along the protofilament decreases by 0.8 Å. Changes in the number of L and R protofilaments govern supercoiling of the filament. Here we report the 2.0 Å resolution crystal structure of a Salmonella flagellin fragment of relative molecular mass 41,300. The crystal contains pairs of antiparallel straight protofilaments with the R-type repeat. By simulated extension of the protofilament model, we have identified possible switch regions responsible for the bi-stable mechanical switch that generates the 0.8 Å difference in repeat distance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Berg, H. C. & Anderson, R. A. Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382 (1973).
Silverman, M. & Simon, M. Flagellar rotation and the mechanism of bacterial motility. Nature 249, 73–74 (1974).
Kudo, S., Magariyama, Y. & Aizawa, S.-I. Abrupt changes in flagellar rotation observed by laser dark-field microscopy. Nature 346, 677–680 (1990).
Ryu, W. S., Berry. R. M. & Berg, H. C. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447 (2000).
Larsen, S. H., Reader, R. W., Kort, E. N., Tso, W. W. & Adler, J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature 249, 74–77 (1974).
Macnab, R. M. & Ornston, M. K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 112, 1–30 (1977).
Turner, L., Ryu, W. S. & Berg, H. C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801 (2000).
O'Brien, E. J. & Bennett, P. M. Structure of straight flagella from a mutant Salmonella. J. Mol. Biol. 70, 133–152 (1972).
Asakura, S. Polymerization of flagellin and polymorphism of flagella. Adv. Biophys. (Japan) 1, 99–155 (1970).
Calladine, C. R. Construction of bacterial flagella. Nature 225, 121–124 (1975).
Calladine, C. R. Design requirements for the construction of bacterial flagella. J. Theor. Biol. 57, 469–489 (1976).
Calladine, C. R. Change of waveform in bacterial flagella: The role of mechanics at the molecular level. J. Mol. Biol. 118, 457–479 (1978).
Kamiya, R. & Asakura, S. Helical transformations of Salmonella flagella in vitro. J. Mol. Biol. 106, 167–186 (1976).
Kamiya, R. & Asakura, S. Flagellar transformations at alkaline pH. J. Mol. Biol. 108, 513–518 (1977).
Hotani, H. Micro-video study of moving bacterial flagellar filaments III. Cyclic transformation induced by mechanical force. J. Mol. Biol. 156, 791–806 (1982).
Kamiya, R., Asakura, S., Wakabayashi, K. & Namba, K. Transition of bacterial flagella from helical to straight forms with different subunit arrangements. J. Mol. Biol. 131, 725–742 (1979).
Yamashita, I. et al. Structure and switching of bacterial flagellar filament studied by X-ray fiber diffraction. Nature Struct. Biol. 5, 125–132 (1998).
Mimori, Y. et al. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J. Mol. Biol. 249, 69–87 (1995).
Morgan, D. G., Owen, C., Melanson, L. A. & DeRosier, D. J. Structure of bacterial flagellar filaments at 11 Å resolution: Packing of the α-helices. J. Mol. Biol. 249, 88–110 (1995).
Mimori-Kiyosue, Y., Vonderviszt, F., Yamashita, I., Fujiyoshi, Y. & Namba, K. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc. Natl Acad. Sci. USA 93, 15108–15113 (1996).
Mimori-Kiyosue, Y., Vonderviszt, F. & Namba, K. Locations of terminal segments of flagellin in the filament structure and their roles in polymerization and polymorphism. J. Mol. Biol. 270, 222–237 (1997).
Mimori-Kiyosue, Y., Yamashita, I., Fujiyoshi, Y., Yamaguchi, S. & Namba, K. Role of the outermost subdomain of Salmonella flagellin in the filament structure revealed by electron cryomicroscopy. J. Mol. Biol. 284, 521–530 (1998).
Samatey, F. A., Imada, K., Vonderviszt, F., Shirakihara, Y. & Namba, K. Crystallization of the F41 fragment of flagellin and data collection from extremely thin crystals. J. Struct. Biol. 132, 106–111 (2000).
Vonderviszt, F., Uedaira, H., Kidokoro, S. -I. & Namba, K. Structural organization of flagellin. J. Mol. Biol. 214, 97–104 (1990).
Honda, S., Uedaira, H., Vonderviszt, F., Kidokoro, S. -I. & Namba, K. Folding energetics of a multidomain protein, flagellin. J. Mol. Biol. 293, 719–732 (1999).
Yoshioka, K., Aizawa, S. -I. & Yamaguchi, S. Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J. Bacteriol. 177, 1090–1093 (1995).
Nogales, E., Sharon, G. W., & Downing, K. H. Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998).
Yamashita, I. et al. Radial mass analysis of the flagellar filament of Salmonella: Implications for subunit folding. J. Mol. Biol. 253, 547–558 (1995).
Namba, K., Yamashita, I. & Vonderviszt, F. Structure of the core and central channel of bacterial flagella. Nature 342, 648–654 (1989).
Vonderviszt, F., Aizawa, S. -I. & Namba, K. Role of the disordered terminal regions of flagellin in filament formation and stability. J. Mol. Biol. 221, 1461–1474 (1991).
Mandelkow, E. -M., Mandelkow, E. & Milligan, R. A. Microtubule dynamics and microtubule caps: A time-resolved cryo-electron microscopy study. J Cell Biol. 114, 977–991 (1991).
Corpet, F., Gouzy, J. & Kahn, D. The ProDom database of protein domain families. Nucleic Acids Res. 26, 323–326 (1998).
Kanto, S., Okino, H., Aizawa, S. -I. & Yamaguchi, S. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J. Mol. Biol. 219, 471–480 (1991).
Kamiya, R., Asakura, S. & Yamaguchi, S. Formation of helical filaments by copolymerization of two types of ‘straight’ flagellins. Nature 286, 628–630 (1980).
Yonekura, K. et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290, 2148–2152 (2000).
Yamamoto, M., Kumasaka, T., Fujisawa, T. & Ueki, T. Trichromatic concept at Spring-8 RIKEN beamline I. J. Synchrotron Rad. 5, 222–225 (1998).
Ueki, T. & Yamamoto, M. The start of a new generation: the present status of the Spring-8 synchrotron and its use in structural biology. Structure 7, R183–R187 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode (Academic, New York, 1997).
Leslie, A. G. W. CCP4/ESF-EACMB. Newslett. Protein Crystallogr. Vol. 26 (Daresbury Laboratory, Warrington, UK, 1992).
Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Terwilliger, T. C. & Berendzen, J. Automated structure solution for MIR and MAD. Acta Crystallogr. D 55, 849–861 (1999).
Brünger, A. T., Kuriyan, J. & Karplus, M. Crystallography R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Bacon, D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524. (1997).
Sayle, R. A. & Milner-White, E. J. RasMol: Biomolecular graphics for all. Trends Biochem. Sci. 20, 374–376 (1995).
Acknowledgements
We thank T. Tomizaki, L. Dumon, W. Burmeister, S. Arzt and S. Wakatsuki at ESRF, and M. Kawamoto, N. Kamiya and K. Miura at SPring-8 for technical help with beamlines. We also thank I. Yamashita and K. Hasegawa for a mutant strain of Salmonella that produces SJW1655-derived site-directed mutant flagellin (G365C), which forms the R-type straight flagellar filament, and helpful information of heavy-atom binding to the filament; J. Tame for critically reading the manuscript; and S. Asakura, T. Nitta and F. Oosawa for continuous support and encouragement.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Samatey, F., Imada, K., Nagashima, S. et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410, 331–337 (2001). https://doi.org/10.1038/35066504
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35066504
This article is cited by
-
Exploiting bacterial-origin immunostimulants for improved vaccination and immunotherapy: current insights and future directions
Cell & Bioscience (2024)
-
Programmable supramolecular chirality in non-equilibrium systems affording a multistate chiroptical switch
Nature Communications (2023)
-
Differences in flaA gene sequences, swimming motility, and biofilm forming ability between clinical and environmental isolates of Aeromonas species
Environmental Science and Pollution Research (2022)
-
Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease
Nature Plants (2020)
-
Identification and characterization of the proteolytic flagellin from the common freshwater bacterium Hylemonella gracilis
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.