Abstract
Patterns of global histone modifications have been recently suggested as outcome predictors in cancer patients. To date, there has been no report on the prognostic significance of global histone modifications in esophageal squamous cell carcinoma. We investigated the role of global histone modification as outcome predictor in patients undergoing esophagectomy for esophageal squamous cell carcinoma. A retrospective clinicopathologic analysis was undertaken of 97 patients with esophageal squamous cell carcinoma who recovered from esophagectomy. Immunohistochemical expression of five histone modification markers, acetylated histone 3 lysine 18 (H3K18Ac), acetylated histone 4 lysine 12 (H4K12Ac), dimethylated histone 4 arginine 3 (H4R3diMe), dimethylated histone 3 lysine 4 (H3 K4diMe), and trimethylated histone 3 lysine 27 (H3K27triMe) was assessed in paraffin-embedded tumor samples. Results were analyzed in relation to patients’ clinicopathologic parameters. There was a positive relationship between tumor differentiation and H3K18Ac (P<0.001), H4R3diMe (P=0.003), and H3K27triMe (P<0.001). Expression of H3K27triMe correlated positively with nodal (N) status (P=0.012) and stage (P=0.025). Univariate analysis showed that better survival in patients with low expression of H3K18Ac (P=0.038) and H3K27triMe (P=0.003). Multivariate analysis showed that nodal status, metastasis status (M), and expression of H3K27triMe predicted survival independently (P<0.001, P=0.016, and 0.048, respectively). Low expression of H3K18Ac and H3K27triMe correlated with better prognosis of patients with esophageal squamous cell carcinoma, especially for those of early stages. We hypothesize that expression of H3K27triMe may be considered as a significant survival predictor for patients with esophageal squamous cell carcinoma.
Similar content being viewed by others
Main
Esophageal cancer accounts for 1–3% of all cancers occurring in the United States and there is a higher incidence in Asia.1 Despite therapeutic advances designed to improve treatment outcome, 5-year survival in patients amenable to definitive treatment remains only 5–30%.2 Currently, the TNM stage remains the most important guide to treatment and is the strongest determinant of prognosis. However, there are substantial differences in survival within stages, probably attributable to differences in biologic behavior of the tumors.3
Epigenetic changes may contribute to the development and progression of cancer and leukemia.4 Epigenetics refers to a number of molecular mechanisms that regulate gene expression without changing the DNA sequence. By far the most studied mechanisms are DNA methylation, covalent modification of histone tails, chromatin remodeling, and microRNAs.5, 6, 7 Epigenetic deregulation may be involved in tumor cell biology, including cell growth, differentiation, and cell death, and therefore may be linked to patient prognosis.8 Methylation of histones at key lysine or arginine residues has been shown to work in concert with acetylation and other modifications to provide a histone code that may determine heritable transcriptional states.4, 9
Two recent reports on global patterns of histone modification suggest their role in outcome for prostate cancer and resected non-small cell lung cancer (NSCLC).10, 11 To date, there has been no report on the prognostic significance of global histone modifications in esophageal squamous cell carcinoma. We hypothesize that histone modification may also play a role in prognosis predication in esophageal squamous cell carcinoma. Employing immunohistochemistry for a panel of previously reported modified histones, the current study was designed to investigate the pattern of global histone modification to see whether there is correlation with prognosis following esophagectomy.
Materials and methods
Case Selection and Tumor Samples
Archival paraffin-embedded tissue blocks were obtained retrospectively of primary tumor specimens from 97 patients who underwent esophagectomy for esophageal squamous cell carcinoma from January 1998 to January 2002. These were consecutive patients who had a complete resection of the tumor and sufficient follow-up data with an exclusion of surgical mortality. Patients included in this study did not receive preoperative or post-operative treatments. Waiver of patient consent was granted by the Institutional Review Board, which allowed us to get access to patients’ medical records and to obtain tissue samples and pertinent follow-up data. From review of medical records, clinical information was collected and TNM status was recorded based upon the American Joint Committee on Cancer staging system12 for esophageal cancer. Survival and follow-up data were obtained from the Institutional Cancer Registry.
Immunohistochemistry for Histone Markers
Paraffin blocks of tumors were cut into 5 μm slices and then processed using standard deparaffinization and rehydration techniques. After antigen retrieval using microwave heating, all tissue sections on slides were immunostained following instructions from the antibody suppliers. A panel of previously reported, highly selective histone antibodies were used as primary antibodies: acetylated (Ac) H3 Lysine (Lys) 18 (K18) (H3K18Ac; rabbit polyclonal, 1:50; Cell Signaling Technology Inc., Danvers, MA, USA), acetylated (Ac) H4 Lys12 (H4K12Ac; rabbit polyclonal, 1:25; Cell Signaling Technology Inc.), dimethylated (diMe) H4 Arg 3 (R3) (H4R3diMe) (rabbit polyclonal, 1:200; Abcam Inc., Cambridge, MA, USA), H3 K4diMe (rabbit polyclonal, 1:200; Abcam Inc.)10, 11 and trimethylated (triMe)H3K27 (H3K27triMe) (mouse monoclonal, 1:200; Upstate Biological, Charlottesville, VA, USA).13 The binding of primary antibody was visualized using a detection system (DAKO LSAB Kit K675; DakoCytomation California Inc., Carpinteria, CA, USA). The normal staining patterns for each of the histones studied were nuclear. Staining results were examined by two observers masked to patients’ clinical information. Another reading by a third observer was needed to reach a consensus when there was a significant discrepancy between initial readings. The percentage of the tumor cells that exhibited a positive immunoreactivity was determined at × 200 magnification using a light microscope (Olympus BX51; Olympus America Inc.). At least 10 high-power fields were examined in each case. The percentage of nuclear staining of tumor cells was calculated10, 11 as close as to the multiples of 10 by the reading pathologists. For correlation with patients’ clincopathologic features and survival, readings for each of the 97 tumor specimens were dichotomized into two categories as low expression (lower than and equal to) and high expression (higher than). Median for the percentage of numbers of positively stained cells in all tumor samples was assessed for each histone antibody. Rabbit serum and mouse isotype-matched IgG served as negative controls for rabbit polyclonal and mouse monoclonal antibody, respectively.
Statistical Analysis
The association between the histone markers and different clinicopathologic characteristics of the patients, including age, gender, tumor differentiation, stage, and TNM category were evaluated by Pearson's χ2 or Fisher's exact test as appropriate. The Kaplan–Meier method14 was used for estimating probability of survival and for univariate analysis. The log-rank test15 was used to assess the significance of difference between pairs of survival probabilities. The Cox proportional hazard model was used to evaluate the association between various histone modifications and patient's survival. For, survival time (measured in months) was the dependent variable. In addition to those five histone markers, we also included stage, age, sex, and differentiation score as potential confounders. Stage (I and II for early stage versus III and IV for late stage), sex, and age (70 and above versus below 70) were treated dichotomously. There were three categories (well, moderate, and poor) in the measure of tumor differentiation. Multivariate analysis for survival was performed using this model. The SPSS 13.0 statistical software package (SPSS Inc., Chicago, IL, USA) was employed for all analysis. A P-value less than 0.05 was considered statistically significant.
Results
There were 92 male and 5 female patients, aged from 38 to 81 years, who underwent esophagectomy for esophageal squamous cell carcinoma and had their clincopathologic features, including differentiation of tumor and staging information, reviewed and summarized in Table 1.
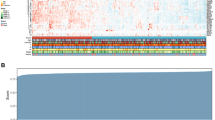
The staining pattern was nuclear in general with similar intensity across all five markers. Representative results of immunohistochemistry for H3K18Ac and H3K27triMe are shown in Figure 1a–f. Percentage of number of cells stained positively for anti-H3K18Ac, -H4K12Ac, -H3K4diMe, -H4R3dime, and -H3K27triMe antibodies ranged from 0 to 100% with a median of 40, 50, 70, 60, and 60%, respectively. The weighted κ-coefficient to reflect agreement in the scoring of immunostaining by different raters for H3K18Ac, H4K12Ac, H3K4diMe, H4R3diMe, and H3K27triMe was 0.858, 0.857, 0.868, 0.887, and 0.891 for H3K27, respectively. Distributions of positively stained tumor cells plotted against their frequencies in patients’ samples examined are shown in Figure 1g. H3K18Ac, H4R3diMe, and H3K27triMe correlated positively with tumor differentiation (Table 2). For other clincopathologic variables, only H3K27triMe showed positive correlation with N status, respectively (Table 2).
Immunohistochemitry for H3K18Ac and H3K27triMe in tumor samples of esophageal squamous cell carcinoma (a–f) and distribution of staining for the five different histone marker antibodies (g). Characteristic nuclear staining is visualized as brown precipitate in the tumor cells. Representative H3K18Ac staining in patients with frequency of positively stained cell of 90, 50, and 20% are shown in (a–c). Similar staining pattern and frequencies of H3K27triMe in tumor cells of esophageal squamous cell carcinoma are shown in (d–f). Distribution of staining frequencies for the five different histone marker antibodies across all 97 tumor samples are shown in (g). The y axis represents the number of patient samples showing positive staining for the indicated percentage of cells stained (x axis).
The overall median survival of this study population was 27 months (95% CI, 19.5–34.5 months). Univariate analysis for clinicopathologic variables and histone modification patterns was performed to determine survival predictor(s) (Table 3). For clinicopathologic variables, nodal status (N), metastasis (M), and tumor stage showed significant correlation with patients survivals. For histone modification patterns, the groups of low expression of H3K18Ac and H3K27triMe showed significantly better survivals than those of high expression (Figures 2 and 3, respectively), whereas other histone markers did not show significant correlation with patients’ survivals (Table 3). Median survival duration in months was significantly different for low and high expression of H3K18Ac and H3K27triMe (Table 3). We were interested in determining whether these two histone modification patterns were early or late event in esophageal squamous cell carcinoma, we further analyzed survivals in relation to expression of H3K18Ac and H3K27triMe between patients with early (I and II) versus late (III and IV) stages. It showed that both of these two histone modifications predict survivals significantly only for patients with early stages (Figures 2 and 3, respectively).
Kaplan–Meier survival curve with log-rank test for 97 patients after resection for esophageal squamous cell carcinoma. Comparison of overall survival, survival for early (I and II) and late (III and IV) (c) stages between patients with low and high expression of H3K18Ac is shown in (a–c), respectively.
Kaplan–Meier survival curve with log-rank test for 97 patients after resection for esophageal squamous cell carcinoma. Comparison of overall survival, survival for early (I and II) and late (III and IV) (c) stages between patients with low and high expression of H3K27triMe is shown in (a–c), respectively.
To determine which of variables analyzed including histone modification patterns was/were independent survival predictor(s), multivariate analysis was performed and showed that expression of H3K27triMe predicted survivals independently (Table 4).
Discussion
Currently, prediction of outcome and guide for therapy for cancer patients is based generally on degree of tumor differentiation and/or tumor stage. It is known that for cancers that are of the same grade and stage, there are subgroups of patients who are molecularly heterogeneous and have different clinical outcomes. It is therefore important to search for reliable molecular biomarkers to better distinguish among those subtypes of patients.3, 4, 16 The concept of molecular staging has been proposed and may be clinically applied to the outcome predication and treatment for lung and esophageal cancer.3, 16 Similar to gene expression, epigenetic alterations have also been used as biomarkers in recent years.17, 18, 19 Relative to DNA methylation, the association between histone modifications and outcome of cancers are less well characterized. Although no promoter-specific histone modifications have been related to cancer prognosis, recent work suggests that global histone modification patterns of the tumor can be used to predict cancer outcome.10, 11 In view of lack of investigation of prognostic significance of global histone modifications in esophageal squamous cell carcinoma, we designed this study to determine whether certain histone modification patterns may serve as outcome predictors for patients undergoing resection for esophageal squamous cell carcinoma.
Histone modification markers employed in this report were selected based on previous studies that correlate patients’ clinical outcome with different histone modification patterns in a variety of cancers.10, 11, 13 Our results demonstrated that all five markers showed a median from 40 to 70% of positively stained cells within the tumor with heterogeneous patterns for distribution of staining frequency. These results indicated that the levels of histone modifications differed considerably QJ;between individual tissues and these differences may be important in defining patient groups of different clinical outcomes.10 Among them, H3K18Ac, H4R3diMe, and H3K27triMe correlated positively with tumor differentiation. These results were in general agreement with results for H3K18Ac and H4R3diMe that were reported by Seligson et al.10 In contrast to theirs that did not include H3K27triMe, we demonstrated a positive correlation between expression of H3K27triMe and tumor differentiation, nodal status and stage as well. Taken together, these results suggest that these three histone modifications are associated with increased gene activity 10 and possibly the aggressiveness of cancer phenotype.
To determine whether the histone modification patterns examined in the current study predicted patients’ clinical outcome, we have shown by univariate Kaplan–Meier survival analysis that H3K18Ac and H3K27triMe correlated significantly with patients’ survival. As a comparison, Seligson et al10 showed that H3K18Ac and H4R3dime predict patients outcome in prostate cancer.20 Our results showing that H3K18Ac correlated with patients survival was similar to theirs but differed in H4R3diMe which did not correlate with patients’ survival. In contrast, Barlesi et al11 report that H2AK5Ac, H3K9Ac, and H4R3diMe are outcome predictors for patients with NSCLC and demonstrated that the frequency of H3K9Ac is higher in adenocarcinoma, whereas that of H4R3diMe is higher in non-adenocarcinoma. Taking these results together, one may speculate that the global histone modification patterns may vary in different cancer types and even in different histologic types of certain tumors.11 To understand further that at early or late stage these correlations were more significant, we did survival analysis to earlier stages (I and II) and later stages (III and IV). These results demonstrated that significant correlations were observed predominately at earlier stages for both of these two modification patterns, suggesting that these molecular events may serve as biomarkers for early detection of esophageal squamous cell carcinoma. Recent studies on histone modification provide novel therapeutic targets in cancer.
Methylation of H3K27 has been implicated in the aggressive phenotype of cancer cells through repression of a panel of tumor suppression genes.21, 22 H3K27 methylation mediated by the primary H3K27 methyltransferase, enhancer of Zeste homolog 2 (EZH2), a component of the Polycomb (PcG) complex that are involved in early carcinogensis.23 As aforementioned, our results showing that H3K27triMe predicted patients’ survival predominately in early stages of esophageal squamous cell carcinoma may in part support the link between PcG complex that leads to H3K27triMe in early carcinogensis.23 Recent studies have shown that EZH2 is overexpressed in a variety of human cancers24 and its expression level correlates with a poor prognosis in prostate24 and breast cancers.25 To date, prognostic significance of expression of its substrate histone protein, H3K27 in cancer is rarely studied. One of the major findings in this study is a significant positive correlation between expression of H3K27triMe and clinicopathologic parameters such as tumor stage and N status (nodal involvement) and it served as a significant survival predictor, supported by multivariate analysis, for patient with esophageal squamous cell carcinoma. One may speculate that increased EZH2 expression may account for high expression of H3K27triMe correlating with poor prognosis in esophageal squamous cell carcinoma.
Schlesinger et al22 employed chromatin immunoprepitation (ChIP) with gene sequencing to investigate the biologic consequence of PcG-mediated H3K27triMe in gene expression across a panel of human cancers and conclude that it functions as a repressor for expression of a wide variety of tumor suppressor genes. The function of PcG-mediated H3K27triMe as a repressor to tumor suppressor genes in breast cancer is further supported by targeting trimethylase of H3K27 using a selective inhibitor, 3-deazaneplanocin A (DZNep).26 In contrast, a more recent study has shown that loss of H3K27triMe correlates with poor prognosis of patients with breast, ovarian, and pancreatic cancer.27 They speculate that contradictory results to those of the EZH2 as described above24, 25 may be attributed to a suppression of certain oncogenes or alternatively a differential regulation of PcG proteins (eg EED or SUZ12) that leads to loss of methylase activity of EZH2.27 Discrepancies in these studies may arise from different types that are studied or a possible differential regulation by H3K27triMe in gene expression. Taken together, these results suggest that biologic significance or clinical implication of H3K27triMe in cancer remains elusive, thus mandating further investigations.
Previous studies show that several histone lysine methyltransferases are linked to cancer.24, 25, 26, 28 The prognostic cellular histone modification patterns may not only be used as general biomarkers but could have specific implications for therapies involving inhibitors to enzymes that catalyze histone modification such as histone deacetylation or methylation.20 If we could verify involvement of these histone methyltransferases, for example by targeting these enzymes by their inhibitors,26 employing in vitro cell line studies or in vivo by animal models29 for esophageal squamous cell carcinoma, one may propose a role for H3K27triMe as a potential pharmacologic therapeutic target for esophageal squamous cell carcinoma. Nevertheless, one should note that there were limitations of this study. First, as we tested the five histone markers in a fashion of multiple comparisons, each time there might be a 5% chance of committing error and the significant results may be obtained by chance alone. Second, our study was based on analysis of a limited number of cases from a single institution. Therefore, one may need to take into account concerns as described above when interpreting these results.
In conclusion, we demonstrated that two histone modifications, H3K18Ac and H3K27triMe, correlated with survivals for patients with esophageal squamous cell carcinoma especially in early stages. We hypothesize that expression of H3K27triMe may be considered as a significant survival predictor for patients with esophageal squamous cell carcinoma. However, this observation may warrant a prospective study of a larger patient cohort to confirm further its role as an independent survival predictor. Hopefully, these results may help us in the prediction of patients’ outcome and in the development of potential target therapy for esophageal squamous cell carcinoma.
References
Layke JC, Lopez PP . Esophageal cancer: a review and update. Am Fam Physician 2006;73:2187–2194.
Tabernero J, Macarulla T, Ramos FJ, Baselga J . Novel targeted therapies in the treatment of gastric and esophageal cancer. Ann Oncol 2005;16:1740–1748.
Lau CL, Moore MB, Brooks KR, D’Amico TA, Harpole Jr DH . Molecular staging of lung and esophageal cancer. Surg Clin North Am 2002;82:497–523.
Lund AH, van Lohuizen M . Epigenetics and cancer. Genes Dev 2004;18:2315–2335.
Gibbons RJ . Histone modifying and chromatin remodelling enzymes in cancer and dysplastic syndromes. Hum Mol Genet 2005;14 (Spec No 1):R85–R92.
Bhalla KN . Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J Clin Oncol 2005;23:3971–3993.
Esteller M . The necessity of a human epigenome project. Carcinogenesis 2006;27:1121–1125.
Jones PA, Baylin SB . The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428.
Jenuwein T, Allis CD . Translating the histone code. Science 2001;293:1074–1080.
Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005;435:1262–1266.
Barlesi F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol 2007;25:4358–4364.
Greene FL . AJCC Cancer Staging Manual 6th edn. American Joint Committee on Cancer, American Cancer Society: New York, 2002;93–94.
Cha TL, Zhou BP, Xia W, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005;310:306–310.
Kaplan ELMP, Meier P . Nonparametric estimation from incomplete observation. J Am Stat Assoc 1958;53:457–481.
The Lifetest Procedure I. SAS Technical Report: P-179, Additional SAS/STAT Procedures, Release 6.03. SAS Institute: Cary, NC, 1988, pp 49–90.
D’Amico TA . Molecular staging and the selection of therapy for non-small cell lung cancer. Semin Thorac Cardiovasc Surg 2005;17:180–185.
Herman JG, Baylin SB . Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042–2054.
Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118–1128.
Esteller M . Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer 2007;96 (Suppl):R26–R30.
Kurdistani SK . Histone modifications as markers of cancer prognosis: a cellular view. Br J Cancer 2007;97:1–5.
Abbosh PH, Montgomery JS, Starkey JA, et al. Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res 2006;66:5582–5591.
Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232–236.
Brock MV, Herman JG, Baylin SB . Cancer as a manifestation of aberrant chromatin structure. Cancer J 2007;13:3–8.
Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624–629.
Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003;100:11606–11611.
Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007;21:1050–1063.
Wei Y, Xia W, Zhang Z, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog 2008;47:701–706.
Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005;121:167–178.
Martin C, Zhang Y . The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005;6:838–849.
Acknowledgements
We thank Dr Mark Ferguson, Professor of Surgery, at the University of Chicago Hospitals for his review of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tzao, C., Tung, HJ., Jin, JS. et al. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod Pathol 22, 252–260 (2009). https://doi.org/10.1038/modpathol.2008.172
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.172
Keywords
This article is cited by
-
KDM6B promotes ESCC cell proliferation and metastasis by facilitating C/EBPβ transcription
BMC Cancer (2021)
-
Histone code and long non-coding RNAs (lncRNAs) aberrations in lung cancer: implications in the therapy response
Clinical Epigenetics (2017)
-
Elevated H3K18 acetylation in airway epithelial cells of asthmatic subjects
Respiratory Research (2015)
-
Interplay among epigenetic alterations and crosstalk between genetic and epigenetic alterations in esophageal squamous cell carcinoma
Esophagus (2014)
-
Epigenetics in esophageal cancers
Cell and Tissue Research (2014)