Abstract
Mucoepidermoid carcinoma is the most common primary malignancy of the salivary gland. We and others showed that CRTC1–MAML2 gene fusion was associated with favorable clinicopathological tumor features. Recently, a novel gene fusion, CRTC3–MAML2, was reported as a rare gene alteration in a case of mucoepidermoid carcinoma. However, its frequency and clinicopathological significance remains unclear. In all, 101 cases of mucoepidermoid carcinoma and 89 cases of non-mucoepidermoid carcinoma of the salivary gland were analyzed, and RNA was extracted from formalin-fixed, paraffin-embedded specimens. In the CRTC family, there have been three genes, CRTC1, CRTC2, and CRTC3. We developed reverse transcription-polymerase chain reaction (RT-PCR) assays for CRTC1–MAML2, CRTC2–MAML2, and CRTC3–MAML2 fusions. Clinicopathological data of the patients were obtained from their clinical records. Of 101 cases of mucoepidermoid carcinoma, 34 (34%) and 6 (6%) were positive for CRTC1–MAML2 and CRTC3–MAML2 fusion transcripts. However, in the 89 cases of non-mucoepidermoid carcinoma, neither transcript was noted. In the former cases, CRTC1–MAML2 and CRTC3–MAML2 fusions were mutually exclusive. The other fusion, CRTC2–MAML2, was not detected. We confirmed that the clinicopathological features of CRTC1–MAML2-positive mucoepidermoid carcinomas indicated an indolent course. CRTC3–MAML2-positive mucoepidermoid carcinomas also had clinicopathologically favorable features; all cases showed a less advanced clinical stage, negative nodal metastasis, no high-grade tumor histology, and no recurrence or tumor-related death after surgical resection of the tumor. It is interesting to note that patients with CRTC3–MAML2-positive tumors (mean 36 years of age) were significantly younger that those with the CRTC1–MAML2 fusion (55 years) and those with fusion-negative tumors (58 years). In conclusion, CRTC3–MAML2 fusion, which is mutually exclusive with CRTC1–MAML2 fusion and specific to mucoepidermoid carcinoma, may be detected more frequently than previously expected. Mucoepidermoid carcinomas possessing CRTC3–MAML2 fusion may be associated with favorable clinicopathological features and patients may be younger than those with CRTC1–MAML2 fusion or those with no detectable gene fusion.
Similar content being viewed by others
Main
Mucoepidermoid carcinoma, representing 5% of all salivary gland tumors and 20% of the malignant forms, is the most frequent primary malignancy of the salivary gland in both adults and children.1 A subset of this carcinoma has been associated with a recurring chromosomal translocation, t(11;19)(q21;p13), which is often the sole cytogenetic alteration.2 This translocation generates a fusion protein comprised of the N-terminal cAMP response element-binding (CREB) protein-binding domain of CREB-regulated transcription coactivator 1 (CRTC1, also called MECT1, TORC1, or WAMP1) at 19q21 and the C-terminal transcriptional activation domain of the Notch coactivator mastermind-like gene 2 (MAML2) at 11q21.3, 4, 5, 6 Recent data suggest that CRTC1–MAML2-induced activation of CREB is critical for cell transformation.3, 5, 6 We recently showed that the CTCR1–MAML2 fusion was detected in approximately 40% of primary salivary gland mucoepidermoid carcinomas, and was associated with a distinct tumor subset that has favorable clinicopathological features and an indolent clinical course.7 Similar observations have been reported by other research groups.8, 9
In the CRTC family, there are two other CRTC1-related human genes, CRTC2 at 1q21 and CRTC3 at 15q26, and the CRTC1 gene has 32% homology with CRTC2 and CRTC3 genes.10 Recently, Fehr et al11 screened 66 mucoepidermoid carcinomas and found that the same part of the MAML2 gene that participates in CRTC1–MAML2 fusion was fused to the CRTC3 gene in a case involving a middle-aged woman. However, the frequency and clinicopathological significance of the CRTC3–MAML2 fusion remains to be clarified. Another gene in the CRTC family, CRTC2, has not been reported to be involved in tumorigenesis in mucoepidermoid carcinoma or other tumors. In this study, we screened a large series of cases of mucoepidermoid carcinoma and non-mucoepidermoid carcinoma for CRTC1–MAML2, CRTC2–MAML2, and CRTC3–MAML2, and studied clinicopathological features of fusion-positive cases.
Materials and methods
Case Selection
Mucoepidermoid carcinomas of the major and minor glands were retrieved from the pathology files of Nagoya City University Graduate School of Medical Sciences; Aichi Cancer Center Central Hospital; Okayama University School of Medicine, Dentistry, and Pharmaceutical Sciences; and Aichi-Gakuin University School of Dentistry. Tumors originating in the lung or other sites were not included in this study. All cases were carefully reviewed according to criteria of the World Health Organization for the classification of head and neck tumors,1 and 101 cases were included in this study. Some of the cases in this series were also included in our previous study.7 Formalin-fixed, paraffin-embedded specimens of the resected tumors were obtained from all cases. In addition, we also collected typical cases of adenoid cystic carcinoma (n=8), oral primary squamous cell carcinoma (n=22), pleomorphic adenoma (n=21), and Warthin's tumor (n=38). Informed consent was obtained, and the study was approved by the institutional review board of Nagoya City University and conducted in accordance with the Declaration of Helsinki. Clinicopathological data were obtained from the medical records. Mucoepidermoid carcinomas were histologically classified according to a three-grade system,1 which has been widely used for grading this carcinoma affecting both major and minor salivary glands. The tumor grade was determined from the sum of the point values assigned to each of five histological elements: cystic component, neural invasion, necrosis, mitosis, and anaplasia (Supplementary 1).
Positive Controls for the CRTC1, CRTC2, and CRTC3 Fusion Transcripts
Clinical samples known to possess the CRTC1–MAML2 fusion were used as the positive control for the CRTC1–MAML2 fusion transcript.7 For positive controls for the other two fusions (CRTC2–MAML2 and CRTC3–MAML2), we synthesized their cDNAs in vitro, as we previously described.12 Figure 1 and Table 1 show the artificial generation of CRTC3–MAML2 cDNA using a series of polymerase chain reaction (PCR) primers. Briefly, a CRTC3-side gene fragment was amplified by the PCR using CRTC3A (20 mer) and a primer A (40 mer) containing both CRTC3 (20 mer) and MAML2 (20 mer) sequences. Similarly, a MAML2-side fragment of the fusion cDNA was amplified by using a primer B (40 mer) containing both 20-mer MAML2 and 20-mer CRTC3 sequences and a MAML2A primer (20 mer). The CRTC3-side and MAML2-side PCR products were diluted to 1:1000, then mixed in a 1:1 ratio, and subjected to PCR using CRTC3A and MAML2A primers. The objective fusion product thus produced was subcloned into pGEM-T Easy Vector (Promega, Madison, WI, USA), and its sequence was confirmed. It was then used as the positive control for CRTC3–MAML2 fusion. Following this strategy, CRTC2–MAML2 cDNA was synthesized and used as a positive control. The breakpoints of CRTC2 and CRTC3 were set so that the exon 1 (CRTC2 and CRTC3) was fused to MAML2 exon 2.7, 11 No CRTC2–MAML2 fusion has been reported, but we assumed that CRTC2 exon 1 would fuse to MAML2 exon 2, based on the homology between CRTC1, CRTC2, and CRTC3 genes and the constant breakpoint of the MAML2 gene when involved in creating the fusion.
Artificial generation of CTCR3–MAML2 fusion cDNA from CRTC3 and MAML2 cDNA. Primer A (40 mer) for CRTC3 cDNA amplification includes 20 mer of the MAML2 sequence on the 3′ side, and primer B (40 mer) for MAML2 cDNA containes 20 mer of the CRTC3 sequence on the 5′ side. After amplification of the CRTC3 gene with primer CRTC3A and primer A and amplification of the MAML2 gene with primer B and MAML2A, both polymerase chain reaction (PCR) products are mixed together, and the fusion gene is amplified by PCR using primers CRTC3A and MAML2A. Arrows indicate breakpoints.
Detection of the CRTC1–, CRTC2–, and CRTC3–MAML2 Fusion Transcripts
CRTC1–,CRTC2–, and CRTC3–MAML2 fusion transcripts were detected using a method consisting of one-tube reverse transcription (RT)-PCR and nested PCR.7 Total RNA was extracted from formalin-fixed, paraffin-embedded specimens as previously described.7 Deparaffinized tissue sections were incubated at 56°C overnight in protease K digestion buffer, and RNA was extracted using concentrated phenol/guanidine isothiocyanate (Trizol LS; Gibco BRL, Friendswood, TX, USA), followed by DNase I treatment (Takara, Otsu, Japan). Then, 5 μl of the extracted RNA were heated to 70°C and placed on ice. The RT-PCR mixture containing outer primers was added. The thermocycler was programmed for an initial RT incubation of 30 min at 42°C and then for 10 min at 95°C for the inactivation of RT as well as for the activation of DNA polymerase. This was followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The products were then diluted 1:50 with water, and subjected to a nested PCR using inner primers. The amplification conditions consisted of 35 cycles at 95°C for 30 s and at 60°C for 30 s. Primers used in this study are shown in Table 1. Primers used for CRTC1–MAML2, CRTC2–MAML2 and CRTC3–MAML2 transcripts were newly designed. As an internal control for RNA quality, the ubiquitously expressed β-actin mRNA fragment (190 bp) was amplified. All specimens were shown to possess RNA of satisfactory quality. The normal salivary gland tissue was used as a negative control.
Statistical Analysis
Statistical evaluation of data from two groups was carried out using the Fischer's exact test and Student's t-test. All analyses were two-tailed. To identify the parameters significantly associated with disease-free and overall survivals, the survival rate was calculated by the Kaplan–Meier method, and the statistical difference was estimated using log-rank test. A value of P<0.05 for each test was regarded as statistically significant, and 0.05 <P<0.1 as marginally significant. All the analyses were performed using the statistical package JMP v5 (SAS Institute, Cary, NC, USA).
Results
Of 101 cases of mucoepidermoid carcinoma, CRTC1–MAML2 and CRTC3–MAML2 fusion transcripts were detected in 34 (34%) and 6 (6%) cases, respectively (Figure 2). These two fusions were mutually exclusive. All fusion transcripts were fused in-frame, and none of the positive cases showed any atypical transcript, such as an insertion or deletion, as confirmed by direct sequencing. The CRTC2–MAML2 fusion transcripts were not detected in any case of mucoepidermoid carcinoma studied. Non-mucoepidermoid carcinoma tumors, including adenoid cystic carcinomas, squamous cell carcinomas, pleomorphic adenomas, and Warthin's tumors were negative for the three fusion transcripts investigated.
Detection of CRTC1–MAML2 fusion (indicated as CRTC1, 93 bp), CRTC2–MAML2 (CRTC2, 85 bp), and CRTC3–MAML2 (CRTC3, 117 bp). P, positive control; N, negative control; bp, base pair; lanes 1–4, tumors positive for CRTC1–MAML2 fusion; lanes 5, 6, tumors positive for CRTC3–MAML2 fusion; lanes 7–10, tumors with no detectable MAML2-associated fusion.
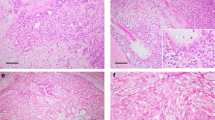
Table 2 shows a summary of the clinical features, treatment, and the outcome of six patients with CRTC3–MAML2-positive mucoepidermoid carcinomas. Two patients were male and four were female patients. The ages ranged from 24 to 53 years (mean 36, median 32). Three tumors were found in the major salivary gland (the parotid), and the remaining three were found in the minor salivary gland (the hard palate, the oral floor, and the retromolar area). Results of laboratory tests including serum lactate dehydrogenase level were unremarkable in all six cases. All tumors were surgically resected without additional treatment, including chemotherapy or radiotherapy. Four tumors were more than 20 mm in diameter. Tumor metastasis to the regional lymph nodes was negative. Histologically, the fusion-positive tumors tended to grow in a solid pattern, and the cystic component was <20% in four cases. No case showed neural invasion, necrosis, or increased mitotic figures. One case showed focal anaplasia (case 2). The histological grade was low in five cases and intermediate in one, with no high-grade tumor (Figure 3). Two patients had clinical stage-I disease and four had stage-II disease. During the follow-up (28–147 months, median 57.5 months), all patients were alive with no evidence of tumor recurrence, and no tumor-related death was recorded.
Table 3 shows a clinicopathological comparison of CRTC3–MAML2 tumors with CRTC1–MAML2 or fusion-negative tumors. As compared with fusion-negative tumors, CRTC3–MAML2 tumors occurred in younger patients (36 years vs 58 years, P=0.0006) and tended to show negative nodal status, a less advanced clinical stage, and lower histological grade (marginally significant). Both CRTC1–MAML2 and CRTC3–MAML2 tumors had favorable clinicopathological features. However, patients with the CRTC3–MAML2 fusion were significantly younger than those with the CRTC1–MAML2 fusion (36 years vs 55 years, P=0.01). Disease-free and overall survival is shown in Figure 4. For disease-free survival, patients with CRTC3–MAML2 and CRTC1–MAML2 fusions showed better prognosis than those with no detectable gene fusion, (P=0.06 and P=0.03, respectively). There was no significant difference in disease-free survival between patients with CRTC3–MAML2 and CRTC1–MAML2 fusions. For overall survival, all patients with CRTC3–MAML2 and CRTC1–MAML2 fusions were alive at the last follow-up. Patients with CRTC3–MAML2 and CRTC1–MAML2 fusions showed longer overall survival than those with no detectable gene fusion (P=0.09 and P=0.0004, respectively).
Disease-free (a) and overall (b) survival of patients with CRTC1–MAML2, CRTC3–MAML2, and no detectable gene fusion. Five-year disease-free survival for patients with CRTC3–MAML2, CRTC1–MAML2, and fusion-negative tumors is 100, 76, and 52%, respectively. Five-year overall survival for patients with CRTC3–MAML2, CRTC1–MAML2, and fusion-negative tumors is 100, 100, and 53%, respectively.
Discussion
In this study, we screened 101 cases of mucoepidermoid carcinoma and 89 cases of non-mucoepidermoid carcinoma tumors for CRTC1–,CRTC2–, and CRTC3–MAML2 fusions using RT-PCR. Positive controls for these assays were either obtained from clinical tumor samples or synthesized in vitro in our laboratory. The CRTC1–MAML2 and CRTC3–MAML2 fusion transcripts were detected in 34 and 6% of mucoepidermoid carcinoma cases, respectively. These two fusions were mutually exclusive. The rate of positives for CRTC3–MAML2 fusion in our series was four times higher than the 1.5% that was reported by Fehr et al11 Several factors that might explain this discrepancy include race (Asian vs Caucasian), geographical factors, case collection bias, and technical differences. Although Fehr et al used conventional RT-PCR, we used nested PCR after RT using gene-specific primers, assuming that our approach may be theoretically more sensitive. The CRTC2 gene is another gene of the CRTC family,10 and we failed to obtain evidence that CRTC2–MAML2 fusion has a role in mucoepidermoid carcinomas or other salivary gland tumors.
It is interesting to note that CRTC3–MAML2 fusion was found only in mucoepidermoid carcinoma cases, but not in non-mucoepidermoid carcinoma tumors. This observation suggests that this fusion may be specific to mucoepidermoid carcinoma, an observation similar to our previous findings as well as those of other groups who showed that CRTC1–MAML2 fusion was found in this carcinoma but not in other salivary tumors.13, 7 Some researchers have reported that the CRTC1–MAML2 fusion was found in a subset of Warthin's tumors,8, 9 whereas others have reported that the fusion occurred only in Warthin's tumors with atypical features.14 All Warthin's tumors that we examined were typical in histology, and no atypical cases were included. Whether atypical Warthin's tumors possess CRTC3–MAML2 fusion may be a subject of future investigation.
Our study showed that mucoepidermoid carcinomas positive for CRTC3–MAML2 fusion had favorable clinicopathological features with no nodal metastasis and a less advanced clinical stage. Although one carcinoma showed focal anaplasia and was given an intermediate grade, five other carcinomas were scored as low grade. It is worth noting that during the follow-up period (median 57.5 months), all patients were alive with no evidence of tumor recurrence after surgery without additional radiation or chemotherapy. These findings suggest that in mucoepidermoid carcinomas positive for CRTC3–MAML2 fusion, extensive surgery may not be necessary, and function-preserving tumor resection may well be warranted. However, we do not exclude a possibility that a subset of CRTC3–MAML2 fusion-positive tumors would progress to high-grade tumors, which may be a heterogeneous group and probably include both de novo high-grade ones and those transformed from low-grade tumors.15 Some high-grade mucoepidermoid carcinomas have been positive for CRTC1–MAML2 fusion.9
In the same subtype of malignant tumor, a difference in the gene partners fused to one pivotal gene can have major clinicopathological implications. In alveolar rhabdomyosarcoma, patients with PAX7–FKHR fusion have a better prognosis than those with PAX3–FKHR fusion.16 Synovial sarcoma patients with tumors positive for SS18–SSX2 fusion show a longer overall survival than those with SS18–SSX1 fusion. In addition, the former tumors are characterized by an exclusively monophasic tumor histology, whereas the latter tumors exhibit a monophasic or biphasic tumor histology.17, 18 In the current study, we presented preliminary data showing that patients with mucoepidermoid carcinoma harboring CRTC3–MAML2 fusion may be younger than those with CRTC1–MAML2 fusion. This finding is important for clarifying mucoepidermoid carcinoma oncogenesis, and further investigation is warranted.
In conclusion, we showed that CRTC3–MAML2 fusion was detected in 6% of mucoepidermoid carcinomas and that the fusion-positive cases showed favorable clinicopathological features and no tumor recurrence after surgical resection alone. It is interesting to note that patients with this gene fusion were comparatively younger than those with the CRTC1–MAML2 fusion or those with no detectable gene fusion. As CRTC3–MAML2 fusion was found only in mucoepidermoid carcinomas, and as all patients with this fusion presented the favorable clinicopathological features and an indolent clinical course, the CRTC3–MAML2 fusion may define a distinct clinicopathological subgroup.
References
Barnes L, Eveson J, Reichart P, et al. editors. Pathology and Genetics of Tumours of the Head and Neck. WHO classification of tumours. IARC Press: Lyon, 2005.
El-Naggar AK, Lovell M, Killary AM, et al. A mucoepidermoid carcinoma of minor salivary gland with t(11;19)(q21;p13.1) as the only karyotypic abnormality. Cancer Genet Cytogenet 1996;87:29–33.
Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 2003;33:208–213.
Enlund F, Behboudi A, Andren Y, et al. Altered Notch signaling resulting from expression of a WAMTP1-2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp Cell Res 2004;292:21–28.
Wu L, Liu J, Gao P, et al. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J 2005;24:2391–2402.
Coxon A, Rozenblum E, Park YS, et al. MECT1- MAML2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res 2005;65:7137–7144.
Okabe M, Miyabe S, Nagatsuka H, et al. MECT1-MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res 2006;12:3902–3907.
Behboudi A, Enlund F, Winnes M, et al. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer 2006;45:470–481.
Tirado Y, Williams MD, Hanna EY, et al. CRTC1/MAML2 fusion transcript in high-grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin's tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer 2007;46:708–715.
Iourgenko V, Zhang W, Mickanin C, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A 2003;100:12147–12152.
Fehr A, Röser K, Heidorn K, et al. A new type of MAML2 fusion in mucoepidermoid carcinoma. Genes Chromosomes Cancer 2008;47:203–206.
Inagaki H, Okabe M, Seto M, et al. API2-MALT1 fusion transcripts involved in mucosa-associated lymphoid tissue lymphoma: multiplex RT-PCR detection using formalin-fixed paraffin-embedded specimens. Am J Pathol 2001;158:699–706.
Martins C, Cavaco B, Tonon G, et al. A study of MECT1-MAML2 inmucoepidermoid carcinoma and Warthin's tumor of salivary glands. J Mol Diagn 2004;6:205–210.
Fehr A, Röser K, Belge G, et al. A closer look at Warthin tumors and the t(11;19). Cancer Genet Cytogenet 2008;180:135–139.
Nagao T, Gaffey TA, Kay PA, et al. Dedifferentiation in low-grade mucoepidermoid carcinoma of the parotid gland. Hum Pathol 2003;34:1068–1072.
Kelly KM, Womer RB, Sorensen PH, et al. Common and variant gene fusions predict distinct clinical phenotypes in rhabdomyosarcoma. J Clin Oncol 1997;15:1831–1836.
Kawai A, Woodruff J, Healey JH, et al. SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998;338:153–160.
Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002;62:135–140.
Acknowledgements
We thank Dr Hitoshi Miyachi and Hideo Fukano, Aichi-Gakuin University School of Dentistry, for their valuable advice. T. Nakayama and S. Miyabe contributed equally to the study and should both be regarded as first authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
Supplementary information
Rights and permissions
About this article
Cite this article
Nakayama, T., Miyabe, S., Okabe, M. et al. Clinicopathological significance of the CRTC3–MAML2 fusion transcript in mucoepidermoid carcinoma. Mod Pathol 22, 1575–1581 (2009). https://doi.org/10.1038/modpathol.2009.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.126
Keywords
This article is cited by
-
The Challenge of “Monomorphic” Mucoepidermoid Carcinoma—Report of a Rare Case with Pure Spindle-Clear Cell Morphology
Head and Neck Pathology (2023)
-
Mucoepidermoid carcinoma of the head and neck: CRTC1/3 MAML 2 translocation and its prognosticators
European Archives of Oto-Rhino-Laryngology (2022)
-
Mucoepidermoid carcinoma of the salivary glands revisited with special reference to histologic grading and CRTC1/3-MAML2 genotyping
Virchows Archiv (2021)
-
Expression of Mucins in Salivary Gland Mucoepidermoid Carcinoma
Head and Neck Pathology (2021)
-
Soft tissue tumor with novel NR1D1-MAML1 fusion in a pediatric case
Virchows Archiv (2020)