Abstract
Myoepithelioma, mixed tumor and parachordoma are uncommon soft tissue tumors thought to represent morphological variants of a single tumor type. The genetic basis of these neoplasms is poorly understood. However, they morphologically resemble mixed tumor of the salivary glands (also known as pleomorphic adenoma), a tumor characterized by deregulated expression of PLAG1 or HMGA2. To evaluate a possible genetic relationship between these soft tissue and salivary gland tumors, PLAG1 expression levels and the genomic status of PLAG1 and HMGA2 were investigated in five soft tissue myoepitheliomas and one pleomorphic adenoma. In addition, all tumors were cytogenetically investigated and whole genome DNA copy number imbalances were studied in five of them. The genetic profiles were heterogeneous and the only aberration common to all soft tissue myoepitheliomas was a minimally deleted region of 3.55 Mb in chromosome band 19p13. Recurrent deletion of CDKN2A suggests that inactivation of this tumor suppressor gene is pathogenetically important in a subset. Furthermore, PLAG1 rearrangement was found in a soft tissue tumor from a patient previously treated for a salivary pleomorphic adenoma, indicating either metastasis of the salivary gland lesion or that some soft tissue tumors develop through the same mechanisms as their salivary gland counterparts.
Similar content being viewed by others
Main
Myoepithelioma of soft tissue is an uncommon and usually benign tumor in which the neoplastic cells display ultrastructural and immunohistochemic features suggestive of myoepithelial differentiation.1 Similar phenotypic characteristics are also found in mixed tumor, a lesion distinguished from myoepithelioma by more pronounced ductal differentiation, and in so-called parachordoma, an ill-defined and controversial tumor that tends to show more prominent cytoplasmic vacuolation.2 Thus, based on the shared morphology with myoepithelial elements, typically within a hyalinized to chondromyxoid stroma, it is increasingly accepted that these three nosologic entities belong to the same family of soft tissue tumors, hereafter referred to as MMP tumors. Clinically, MMP tumors usually present as painless lesions in the extremities of adults but other locations as well as occurrence in the pediatric setting have been described. Apart from the rare cases displaying overt malignant histological features, local recurrences and metastases are uncommon.1
Whereas little is known about the genetics of soft tissue MMP tumors,2, 3, 4, 5, 6, 7 morphologically similar lesions at other sites, notably mixed tumors (also known as pleomorphic adenomas) of the salivary glands, have been more extensively analyzed by cytogenetic and molecular genetic techniques. Most pleomorphic adenomas have a near-diploid chromosome number and two distinct genetic subgroups with recurrent structural rearrangements have been discerned.8 A majority of the cases with abnormal karyotypes have rearrangements affecting chromosome band 8q12, resulting in activation of PLAG1 expression through exchange of promoter sequences.9 In close to 10% of the cases, chromosome region 12q13–15 is affected instead, resulting in deregulated expression of the HMGA2 gene.8 The remaining cases show other rearrangements of unknown molecular significance or normal karyotypes.
Herein, five soft tissue MMP tumors as well as (purely for purpose of comparison) one pleomorphic adenoma of the parotid gland were cytogenetically investigated, and in five of them genomic imbalances were studied using tiling resolution array comparative genomic hybridization (array CGH). To search for possible similarities between soft tissue MMP tumors and pleomorphic adenoma of the salivary glands, PLAG1 expression was analyzed with real-time quantitative PCR (RQ-PCR) and the status of the PLAG1 and HMGA2 loci was investigated with fluorescence in situ hybridization (FISH). Recent data show that rearrangements of the EWSR1 gene could be involved in the development of MMP tumors, specifically through the creation of an EWSR1–PBX1 fusion gene.10 Therefore, also genomic rearrangement of EWSR1 and the presence of an EWSR1–PBX1 fusion gene were investigated.
Materials and methods
Tumor Samples
The study included five patients (cases 1–5) with deep-seated intra- or intermuscular soft tissue MMP tumors located in the thigh, lower leg, lower back and lower arm, respectively, of two men and three women, age range 11–77 years. Two tumors recurred locally, and two metastasized to the lungs. The morphological details of case 3 have been discussed previously.11 Case 4 had a previous history of a pleomorphic adenoma of the parotid gland; the primary tumor and a local recurrence had been excised 12 and 1.5 years, respectively, prior to the diagnosis of the soft tissue tumor. At 1 year after treatment for the soft tissue tumor, this patient was diagnosed with invasive lobular mammary carcinoma. Case 6, a pleomorphic adenoma located in the parotid gland of an 87-year-old woman, was included in the study for comparison. Clinical data are summarized in Table 1.
The present cases represented all tumors diagnosed as an MMP tumor, and from which material for genetic analyses was available, among the approximately 2200 soft tissue tumors analyzed at the Department of Clinical Genetics in Lund, Sweden. In addition, one cytogenetically analyzed case from the Department of Genetics in Porto, Portugal was included. All cases were histologically reviewed by two of the authors (CDMF and HAD).
Chromosome Banding Analysis
Fresh tumor samples were processed for G-banding analysis as previously outlined and karyotypes were described according to the guidelines in ISCN 1995.12, 13
Array Comparative Genomic Hybridization
All tumors except case 4 were analyzed with array CGH using more than 32 000 partly overlapping, individual bacterial artificial chromosome (BAC) clones generating complete coverage of the human genome. The arrays were produced at the Swegene DNA Microarray Resource Center, Department of Oncology, Lund University (http://swegene.onk.lu.se), as previously described,14 using BAC clones mapped to the hg17 genome build. Extraction, labeling and hybridization of genomic DNA from freshly frozen tumor biopsies, as well as pretreatment and washing of slides were performed as described.15 As a control for normal copy number, a DNA pool derived from multiple healthy male donors was used (Promega, Madison, WI, USA). Image and data analyses were performed as described.16 In brief, following background correction the log(2) ratios were calculated for each spot, and unreliable features and spots not showing signal-to-noise ratios ⩾3 for both channels were eliminated. Normalization of data was performed using Lowess normalization and subsequently single outlier probes were removed. Thereafter, the log(2) ratios for each sample and platform was segmented and segments less than 500 kb in size were removed. Copy number alterations were determined by comparing the segmented log(2) ratios to gain/loss thresholds obtained by an adaptive scaling method.17 Segments above gain threshold were set to 1, below loss threshold to −1 and in-between to 0. Segmented log(2) ratios above five times the threshold were defined as amplified, ratios above eight times the threshold were defined as highly amplified and ratios less than seven times the negative threshold were defined as homozygously deleted. Array CGH data are available at GEO (http://www.ncbi.nlm.nih.gov/geo), using the accession number GSE10266.
Fluorescence In Situ Hybridization
FISH analyses were performed as described,18 using the BAC clones RP11-299L9 and RP11-427K2 for HMGA2 and RP11-246A9, RP11-140I16 and RP11-1130K23 for PLAG1 (BACPAC Resources center) on metaphase spreads from cases 2 (only HMGA2) and 4–6. Prior to analysis, the location of the probes was verified by hybridization to normal metaphase cells. The status of the EWSR1 locus was investigated using a break-apart probe (Vysis, Downers Grove, USA). In cases 2, 4 and 5, 125 tumor interphase nuclei were analyzed. In addition, nine abnormal metaphase spreads were analyzed in case 4. Split signals or rearrangement of probes occurring in less than 10% were not considered significant. Whole chromosome paint (WCP) probes specific for selected chromosomes were used to discriminate normal cells from tumor cells (Vysis).
Real-Time Quantitative PCR
RNA was extracted using the guanidine-thiocyanate method,19 and reversely transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The relative expression of PLAG1 was investigated in cases 1–3 and 5–6 using RQ-PCR with the TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). To determine the baseline expression of PLAG1, which is normally not expressed in adult tissues, RNA from normal kidney and lymphocytes from healthy blood donors were used. RNA from a salivary gland pleomorphic adenoma with a known t(3;8)(p21;q12) resulting in promoter swapping between PLAG1 and CTNNB1 was used as a positive control for PLAG1 expression (kindly provided by Dr Carmo Martins, The Portuguese Oncology Institute, Lisbon, Portugal). To correct the expression data for differences in cellular input, RNA quality and reverse transcriptase efficiency, the expression of the housekeeping gene glucuronidase-β (GUSB) was quantified in the same reaction for all samples. Both genes were analyzed with primers and probes located in their 3′-ends, investigating the exon boundaries 4–5 and 11–12 of PLAG1 (NM_002655.1) and GUSB (NM_000181.1), respectively (Applied Biosystems). Calculations were performed using the relative standard curve method, serial dilutions of the positive control were used to construct a calibration curve, and all reactions were performed in triplicate and assayed on an ABI Prism 7000 sequence detection system (Applied Biosystems).
Reverse Transcriptase PCR
Reverse transcriptase PCR (RT–PCR) for the candidate fusion genes EWSR1–PBX1, FUS–PBX1 and TAF15–PBX1 was performed in cases 1–3 and 6 as described,20 using the primer combinations EWS501F-PBX1413R, TLS165F-PBX1372R, TAF308F-PBX1372R and EWS225F-PBX1372R, EWS225F-PBX1064R, TLS294F-PBX1064R, TAF348F-PBX1064R for outer and inner RT–PCR, respectively. Primer sequences are presented in Table 2.
Results
Histological Features
Each of the five soft tissue tumors shared broadly similar cytoarchitectural features and all tumors fulfilled recently defined criteria for soft tissue myoepithelial neoplasms (Figure 1).1 Each tumor was composed predominantly of spindled or more ovoid cells with variably palely eosinophilic or clear cytoplasm and uniform ovoid nuclei showing an evenly distributed chromatin pattern. Cells with clear cytoplasm predominated in case 1 and in one tumor showing strikingly vacuolated (‘parachordoma’-like) cells (case 2). Tumor cells were arranged in trabeculae, nests or fascicles or, focally, in a more sheet-like pattern. The stroma was variably myxoid or hyaline, being predominantly hyaline in case 1. In two cases there was evident ductal differentiation, in which rounded or more stellate ductal structures were lined by more cuboidal epithelial cells. These structures were numerous in one soft tissue lesion from the thigh (case 4) and were identified in just one very small microscopic focus in the salivary gland lesion (case 6), underlining the morphological continuum between myoepithelioma and so-called mixed tumor. None of the cases showed pleomorphism, prominent nucleoli or more than occasional mitoses or necrosis—thus none showed overt features of malignancy as defined in soft tissue myoepithelioma.1 However, the two tumors which behaved in a malignant fashion each showed a striking and unusually infiltrative growth pattern (through fascia/muscle in case 3 and into bone in case 5).
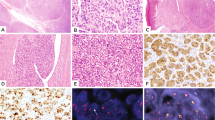
Histologic and immunohistochemic features of soft tissue MMP tumors. (a) Case 1 showing variably eosinophilic or clear cytoplasm and uniform ovoid-to-tapering nuclei. (b) Case 4 showing focally prominent ductal differentiation, embedded in a myxoid matrix (‘mixed tumor’). (c) Case 5 showing uniform cytomorphology with a mixed fascicular and trabecular architecture. (d) Immunohistochemic staining showing positivity for keratin AE1/AE3 in a trabecular clear cell area of case 1. (e) S-100 protein expression in the salivary gland lesion (case 6).
The results of immunohistochemical staining are summarized in Table 3. All cases showed positivity for at least one keratin, 4/4 stained for EMA, 4/6 for S-100 protein, 2/5 for GFAP, 2/2 for calponin, 3/3 for desmin and 1/1 for SMA. The two tumors (cases 1 and 3) that were negative for both S-100 and GFAP showed classical myoepithelial morphology and were positive for at least one myogenic marker.
Cytogenetic Findings
No consistent cytogenetic aberration was seen in the five soft tissue MMP tumors, all of which showed abnormal karyotypes in short-term cultured cells (Table 1). Four tumors had chromosome numbers in the near-diploid range and one was hypotriploid. A primary lesion and its local recurrence surgically excised 8 years later displayed almost identical karyotypes. A deletion in the proximal part of chromosome arm 8q was detected in two cases (cases 4 and 5). The salivary gland pleomorphic adenoma showed an abnormal pseudodiploid karyotype including the characteristic t(3;8)(p21;q12).
Copy Number Alterations
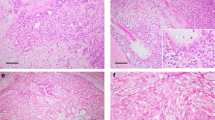
Aberrant DNA copy numbers were found in all five cases studied (Table 4; Figure 2a), and imbalances detected in three or more cases included loss of material at positions 16.42–16.87 Mb in 1p36 (this region harbors known copy number variation, CNV), 19.43–23.18 Mb in 9p21-22, 128.93–138.39 Mb in 9q34, 25.86–114.12 Mb in 13q12-34, 0.02–19.19 Mb in 19p13, 54.64–55.72 Mb in 19q13 (CNV) and gain of the region at position 4.11–6.22 Mb in 5p15. The region at 1.27–4.82 Mb in 19p13 was deleted in all cases. Homozygous deletions were found at positions 19.43–28.24 Mb in 9p21–22 (case 2) and 8.57–8.83 Mb in 19p13 (CNV) (case 6). Case 2 displayed amplification of material from chromosomes 6, 9 and 13 (Figure 2b), located in marker chromosomes (Figure 2c). The salivary gland pleomorphic adenoma (case 6) displayed a 14 Mb deletion in 8q12–13 with a break in, or close to, the 5′ end of PLAG1.
Genomic imbalances and gene rearrangement in soft tissue MMP tumors. (a) The pattern of genomic imbalances in four soft tissue MMP tumors and one pleomorphic adenoma (case 6) was heterogeneous and few recurrent aberrations were found. The individual cases are arranged in rows and the chromosomes in columns, separated by bars. Amplifications, gains and losses are shown in red, dark red and green, respectively. The genomic positions of the imbalances are presented in Table 4. (b) Case 2 showed amplifications of regions on chromosomes 6, 9 and 13 upon array CGH analysis. The individual bacterial artificial chromosome (BAC) clones are arranged according to their respective tumor/reference log 2 ratios on the y axis and their genomic location on the x axis. (c) The amplified material was found to be located in marker chromosomes, as shown by hybridization with whole chromosome paint (WCP) probes for chromosomes 6 (red), 9 (green) and 13 (blue). (d–f) Fluorescence in situ hybridization (FISH) analysis of gene rearrangements was performed using probes (indicated in red, green and blue) covering or partially covering the respective genes (indicated in yellow). In addition, WCP probes specific for selected chromosomes were used to discriminate normal cells from tumor cells. (d) EWSR1 on chromosome 22 was investigated using an EWSR1 break-apart probe (red and green). Case 5 displayed intact signals from this probe in 125 tumor nuclei, detected by three chromosomes 12 (green). (e) The HMGA2 gene on chromosome 12 was analyzed using the probes RP11-299L9 (green) and RP11-427K2 (red), respectively. In case 4, tumor cells identified by a WCP probe for chromosome 3 (green) showed a normal HMGA2 hybridization pattern. (f) Rearrangement of PLAG1 on chromosome 8 was studied using three, partially overlapping probes; RP11-246A9 (green), RP11-140I16 (red) and RP11-1130K23 (blue). In case 4, the PLAG1 gene was rearranged. The 5′ part of PLAG1 and regions upstream of the gene (blue) were translocated to a derivative chromosome 3. Chromosome 3 was detected by a red WCP probe (not shown) and therefore, the red signal on the derivative chromosome 8 possibly represented material from chromosome 3.
FISH Analysis of PLAG1, HMGA2 and EWSR1
Investigation of the genomic status of PLAG1, HMGA2 and EWSR1 showed rearrangement of PLAG1 in cases 4 and 6 and loss in case 5 (Table 1, Figures 2d–f). The other two genes were unaffected.
Expression Levels of PLAG1
The expression level of PLAG1 was seven times higher in case 6 compared with the positive control. In contrast, in the negative controls and the rest of the samples the expression levels of PLAG1 were 8–100 times lower than in the positive control. Due to lack of material, case 4 could not be analyzed.
Fusion Gene Analysis
Neither EWSR1–PBX1 nor the putative fusion transcripts FUS–PBX1 and TAF15–PBX1 were detected (data not shown).
Discussion
Prompted by the morphological similarities between soft tissue MMP tumors and pleomorphic adenoma of salivary glands, a possible genetic relationship between these tumor types was investigated. The most frequently encountered aberration in pleomorphic adenoma is a t(3;8)(p21;q12), resulting in a CTNNB1–PLAG1 gene fusion.21 The pleomorphic adenoma of the present study also displayed this translocation, with a concurrent deletion of the genomic region upstream of PLAG1, as well as increased expression levels of this gene. Rearrangement of PLAG1 was also detected by FISH in one of the soft tissue MMP tumors (case 4). This patient had previously been treated twice for pleomorphic adenoma of the parotid gland, and it can be noted that the soft tissue tumor of this patient shared many histological features with the pleomorphic adenoma investigated here, including prominent ductal differentiation and identical immunohistochemical staining. Unfortunately, neither RNA from the soft tissue tumor nor slides from the salivary gland tumors of case 4 were available. Thus, expression levels of PLAG1 could not be confirmed due to lack of material. The genetic and histological features of the MMP tumor of case 4 could indicate that a subset of MMP tumors indeed develop through the same molecular pathways as their salivary gland counterparts. Another possibility would be that there was a more direct association between the soft tissue MMP tumor and the preexisting pleomorphic adenoma of case 4, ie that the former was a metastasis from the latter. It is well recognized that other tumors may evolve from salivary pleomorphic adenoma, commonest among which is carcinoma ex pleomorphic adenoma but other include carcinosarcoma (‘malignant mixed tumor’), rarely pure sarcoma and perhaps even myoepithelioma.22 Among these, carcinoma ex pleomorphic adenomas may still demonstrate PLAG1 alterations.23 Finally, although an exceedingly rare phenomenon, pleomorphic adenoma can metastasize and give rise to a so-called benign metastasizing pleomorphic adenoma.24 However, none of the reported cases has metastasized to the soft tissues. Reflected by its rareness, little is known about the genetic background of this disease.25 Thus, although it might seem unlikely that a pleomorphic adenoma would give rise to a single soft tissue metastasis, we believe that this scenario is plausible in case 4. A hypothesis previously put forward has suggested that the hematogenous spread of tumor cells in benign metastasizing pleomorphic adenoma may be caused by the surgical manipulation of the primary tumor. Albeit this is only a hypothesis, it could explain the spread of an apparently benign tumor. Furthermore, pleomorphic adenomas have previously shown single metastases in various tissues and although most patients have subsequently presented multiple metastatic locations, the time interval may be several years.24 In this context it could be interesting to note that the patient was diagnosed with breast cancer 1 year after the detection of the soft tissue tumor. Unfortunately, a possible morphological or genetic relationship between these tumors could not be investigated due to lack of tumor material.
The remaining cases neither displayed rearrangements nor altered expression levels of any of the genes investigated. In several bone and soft tissue tumors EWSR1, FUS and TAF15 have been shown to be interchangeable 5′-partners in fusions with various transcription factor encoding genes.26 In this study, we did not find fusion of any of these genes to PBX1. Thus, the potential impact of the EWSR1–PBX1 fusion gene on soft tissue MMP tumor development needs to be evaluated in larger tumor series.
This is the first study characterizing whole genome DNA copy number imbalances in soft tissue MMP tumors and the genomic imbalances detected were diverse. The only recurrent aberration of known importance for tumor development was homo- and heterozygous deletions of the region in 9p21–22 that harbors the CDKN2A and CDKN2B genes. Although CDKN2A is commonly deleted in various tumors, its status in soft tissue MMP tumors is not known. Investigations of this gene in pleomorphic adenoma have shown conflicting results.27, 28 Collectively, however, previous studies suggest that genes involved in the RB1 pathway are likely targets in the development of pleomorphic adenoma. The present findings thus imply that the RB1 pathway is likely to be involved also in the development of soft tissue MMP tumors. Defined by the deletion in case 3, the region 1.27–4.82 Mb on chromosome 19 was heterozygously lost in all cases, including the pleomorphic adenoma. Although this region contains many genes, some of which are involved in the regulation of the cell cycle, growth and apoptosis, there is no obvious candidate to be targeted by this deletion. A complex pattern of aberrations was seen in the primary tumor of case 2 with amplifications, interrupted by normal copy numbers and losses, of regions on chromosomes 6, 9 and 13. Interestingly, the complicated genetic rearrangements found already at the primary excision seemed to persist in the recurrent tumor, excised 8 years later, as the cytogenetic findings were highly similar in the two specimens.
There were several discrepancies between the karyotypes and the array-CGH data, notably in case 2. In part, this is most probably due to the higher resolution of array CGH and to the fact that many aberrations remain unresolved after G-banding analysis. Furthermore, it cannot be excluded that there were additional cell populations that due to poor proliferation in vitro were missed by the cytogenetic analysis.
In conclusion, the heterogeneous genetic findings in an admittedly small series of soft tissue MMP tumors do not indicate that these neoplasms share a common genetic profile, visible at the chromosome or DNA level. Whether this is due to intrinsic heterogeneity of soft tissue MMP tumors or whether the tools to separate phenotypic subgroups within this family of tumors are still too blunt remains to be elucidated.
References
Hornick JL, Fletcher CDM . Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol 2003;27:1183–1196.
Kilpatrick SE, Limon J . Mixed tumour/Myoepithelioma/Parachordoma. In: Fletcher CDM, Unni KK, Mertens F (eds). Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, 2002;198–199.
van den Berg E, Zorgdrager H, Hoekstra HJ, et al. Cytogenetics of a soft tissue malignant myoepithelioma. Cancer Genet Cytogenet 2004;151:87–89.
Pauwels P, Dal Cin P, Roumen R, et al. Intramuscular mixed tumour with clonal chromosomal changes. Virchows Arch 1999;434:167–171.
Folpe AL, Agoff SN, Willis J, et al. Parachordoma is immunohistochemically and cytogenetically distinct from axial chordoma and extraskeletal myxoid chondrosarcoma. Am J Surg Pathol 1999;23:1059–1067.
Limon J, Babinska M, Denis A, et al. Parachordoma: a rare sarcoma with clonal chromosomal changes. Cancer Genet Cytogenet 1998;102:78–80.
Tihy F, Scott P, Russo P, et al. Cytogenetic analysis of a parachordoma. Cancer Genet Cytogenet 1998;105:14–19.
Stenman G . Fusion oncogenes and tumor type specificity—insights from salivary gland tumors. Semin Cancer Biol 2005;15:224–235.
Persson F, Winnes M, Andrén Y, et al. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene 2008;27:3072–3080.
Brandal P, Panagopoulos I, Bjerkehagen B, et al. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1–PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer 2008;47:558–564.
Gleason BC, Fletcher CDM . Myoepithelial carcinoma of soft tissue in children: an aggressive neoplasm analyzed in a series of 29 cases. Am J Surg Pathol 2007;31:1813–1824.
Mandahl N . Methods in solid tumor cytogenetics. In: Rooney DE (ed). Human Cytogenetics: Malignancy and Acquired Abnormalities, 3rd edn. Oxford University Press: New York, 2001, pp 165–203.
Mitelman F . ISCN (1995): An International System for Human Cytogenetic Nomenclature. S Karger: Basel, 1995.
Jönsson G, Staaf J, Olsson E, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer 2007;46:543–558.
Heidenblad M, Hallor KH, Staaf J, et al. Genomic profiling of bone and soft tissue tumors with supernumerary ring chromosomes using tiling resolution bacterial artificial chromosome microarrays. Oncogene 2006;25:7106–7116.
Hallor KH, Staaf J, Jönsson G, et al. Frequent deletion of the CDKN2A locus in chordoma: analysis of chromosomal imbalances using array comparative genomic hybridisation. Br J Cancer 2008;98:434–442.
Staaf J, Jönsson G, Ringner M, et al. Normalization of array-CGH data: influence of copy number imbalances. BMC Genomics 2007;8:382.
Dahlén A, Debiec-Rychter M, Pedeutour F, et al. Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int J Cancer 2003;103:616–623.
Chomzynski P, Sacchi N . Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987;162:156–159.
Hallor KH, Mertens F, Jin Y, et al. Fusion of the EWSR1 and ATF1 genes without expression of the MITF-M transcript in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer 2005;44:97–102.
Kas K, Voz ML, Roijer E, et al. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet 1997;15:170–174.
Yoshizaki T, Himi Y, Minato H, et al. Malignant myoepithelioma arising from recurrent pleomorphic adenoma of minor salivary gland. Auris Nasus Larynx 2002;29:91–94.
Martins C, Fonseca I, Roque L, et al. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol 2005;18:1048–1055.
Marioni G, Marino F, Stramare R, et al. Benign metastasizing pleomorphic adenoma of the parotid gland: a clinicopathologic puzzle. Head Neck 2003;25:1071–1076.
Jin Y, Jin C, Arheden K, et al. Unbalanced chromosomal rearrangements in a metastasizing salivary gland tumor with benign histology. Cancer Genet Cytogenet 1998;102:59–64.
Riggi N, Cironi L, Suvà ML, et al. Sarcomas: genetics, signalling, and cellular origins. Part 1: the fellowship of TET. J Pathol 2007;213:4–20.
Weber A, Langhanki L, Schütz A, et al. Alterations of the INK4a-ARF gene locus in pleomorphic adenoma of the parotid gland. J Pathol 2002;198:326–334.
Patel RS, Rose B, Bawdon H, et al. Cyclin D1 and p16 expression in pleomorphic adenoma and carcinoma ex pleomorphic adenoma of the parotid gland. Histopathology 2007;51:691–696.
Acknowledgements
Carmo Martins is gratefully acknowledged for providing the positive control sample used in the RQ-PCR analysis. This work was supported by the Swedish Cancer Society and the Swedish Childhood Cancer Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
There is no conflict of interest to declare.
Rights and permissions
About this article
Cite this article
Hallor, K., Teixeira, M., Fletcher, C. et al. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol 21, 1311–1319 (2008). https://doi.org/10.1038/modpathol.2008.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.124
Keywords
This article is cited by
-
Soft Tissue Special Issue: Myoepithelial Neoplasms of Soft Tissue: An Updated Review with Emphasis on Diagnostic Considerations in the Head and Neck
Head and Neck Pathology (2020)
-
Multi-dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions
Nature Communications (2017)
-
Histopathological, immunohistochemical and molecular spectrum of myoepithelial tumours of soft tissues
Virchows Archiv (2012)
-
NR4A3 rearrangement reliably distinguishes between the clinicopathologically overlapping entities myoepithelial carcinoma of soft tissue and cellular extraskeletal myxoid chondrosarcoma
Virchows Archiv (2012)