Abstract
Synovial Sarcoma consistently harbors t(X;18) resulting in SS18-SSX1, SS18-SSX2 and rarely SS18-SSX4 fusion transcripts. Of 328 cases included in our study, synovial sarcoma was either the primary diagnosis or was very high in the differential diagnosis in 134 cases: of these, amplifiable cDNA was obtained from 131. SS18-SSX fusion products were found in 126 (96%) cases (74 SS18-SSX1, 52 SS18-SSX2), using quantitative and 120 by conventional reverse transcriptase–polymerase chain reaction (RT-PCR). One hundred and one cases in a tissue microarray, analyzed by fluorescence in situ hybridization (FISH), revealed that 87 (86%) showed SS18 rearrangement: four RT-PCR positive cases, reported as negative for FISH, showed loss of one spectrum green signal, and 15 cases had multiple copies of the SS18 gene: both findings are potentially problematic when interpreting results. One of three cases, not analyzed by RT-PCR reaction owing to poor quality RNA, was positive by FISH. SS18-SSX1 was present in 56 monophasic and 18 biphasic synovial sarcoma: SS18-SSX2 was detected in 41 monophasic and 11 biphasic synovial sarcoma. Poorly differentiated areas were identified in 44 cases (31%). There was no statistically significant association between biphasic, monophasic and fusion type. Five cases were negative for SS18 rearrangement by all methods, three of which were pleural-sited neoplasms. Following clinical input, a diagnosis of mesothelioma was favored in one case, a sarcoma, not otherwise specified in another and a solitary fibrous tumor in the third case. The possibility of a malignant peripheral nerve sheath tumor could not be excluded in the other two cases. We concluded that the employment of a combination of molecular approaches is a powerful aid to diagnosing synovial sarcoma giving at least 96% sensitivity and 100% specificity but results must be interpreted in the light of other modalities such as clinical findings and immunohistochemical data.
Similar content being viewed by others
Main
Synovial sarcoma is an aggressive soft tissue tumor that accounts for 5–14% of soft tissue sarcomas and classically occurs in the extremities of young adults.1, 2, 3, 4, 5 As it is one of the most common soft tissue sarcoma and has been described in a wide variety of anatomical locations and in all ages the diagnosis needs to be entertained frequently.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 The detection of a reciprocal translocation between chromosomes X and 18 t(X;18)(p11.2;q11.2)16, 17, 18 led to the identification of an SS18 gene (also known as SYT), rearrangement being involved in the formation of a SS18-SSX fusion protein in synovial sarcoma.19, 20 SS18-SSX1 and SS18-SSX2 fusions have been detected in approximately 61 and 37% of cases respectively, with the latter being associated with a better overall survival.21 In the vast majority of synovial sarcoma, fusion of SS18 with SSX1 and SSX2 involves the same codons: namely codon 410 on SS18 (ENSG00000141380) fuses with codon 111 on either SSX1 (ENSG00000126752) or SSX2 (ENSG00000157950). Variant chimeric transcripts are rare and have been described only in isolated cases. To our knowledge, these include five variants involving the fusion site of SS18-SSX1,22, 23, 24, 25, 26 and another five involving the SS18-SSX2 fusion site.20, 22, 27, 28, 29 SS18 also rarely forms a fusion gene with SSX4 involving the ‘usual’ SS18-SSX site at codon 410 SS18 and codon in 111 SSX4:30, 31, 32 in addition to this ‘usual type’, two variants have been detected.33, 34 One case of synovial sarcoma has been reported to harbor a fusion involving a gene on chromosome 20 -SS18LI, a homolog of the SS18 gene and the SSX1 gene.35 In addition, Sonobe et al36 reported a variant, which harbored a truncated cDNA, but involved the usual fusion in SS18-SSX1. The function of the SS18-SSX chimeric protein and its relationship to tumor development remains unknown, as does the mechanism through which the translocation occurs.37
There are two major morphological subtypes of synovial sarcoma: the more common monophasic type, representing 60–75% of this tumor,5, 38, 39, 40, 41 is composed of short fascicles of plump spindle cells often with epithelioid areas.4, 38 Identification of well-formed glandular structures admixed with plump spindle and epithelioid cells are required before making the diagnosis of the biphasic type, and although this diagnosis is less problematic this variant shares microscopic features with mesothelioma, mixed mullerian tumors and adenocarcinomas, which have a marked desmoplastic stromal response. Both subtypes may harbor a poorly differentiated component41, 42 and if this area were biopsied, the differential diagnosis includes a wide variety of neoplasms including ‘small round blue cell’ tumors,10 malignant peripheral nerve sheath tumor and non-sarcomatous neoplasms.43, 44 This is particularly the case when a synovial sarcoma occurs at an unusual site such as lung or bone.5, 14 In most cases, immunohistochemistry provides the information necessary for distinguishing these neoplasms. However, there are situations when even immunohistochemistry cannot resolve the diagnostic problem, and in these genetic analysis for the detection of SS18 gene rearrangement has been found to be a powerful diagnostic tool for making the diagnosis of synovial sarcoma.5, 14, 45, 46, 47, 48, 49
There is evidence to suggest that the type of SS18-SSX fusion is associated with the histological subtype.21, 37, 50 The largest study that has addressed this issue is a multi-institutional analysis of 61 biphasic synovial sarcoma of which only three harbored the SS18-SSX2 transcript.21 Although this finding has been reproduced in smaller series,37, 50 others have reported that the existence of a SS18-SSX2 in the biphasic variant may not be such a rare occurrence.27, 37 In addition, data on the type of SS18-SSX fusion in synovial sarcoma with poorly differentiated areas are scant.39
The primary aim of this study was to analyze a large series of formalin-fixed paraffin-embedded soft tissue lesions for the SS18-SSX fusion transcripts and compare three different molecular techniques, so that the findings could be applied to a diagnostic service. We also correlated the SS18-SSX fusion type with the histological subtype and the presence of poorly differentiated areas.
Materials and methods
Selection of Cases
Three hundred and twenty-eight cases were analyzed for the presence of SS18-SSX fusion transcripts. These included 134 cases, 76 resection specimens and 58 biopsies, where synovial sarcoma was either considered the primary diagnosis or where synovial sarcoma was included as part of the main differential diagnosis based on clinical information, morphology and immunohistochemistry. Of these 134 cases, 73 and 50 cases were retrieved from the archives of the Histopathology Departments of the Royal National Orthopaedic Hospital, and Santa Casa de Sao Paulo Central Hospital, Brazil, respectively. Eleven were referred from other UK hospitals. Normal tissue and tumors considered unlikely to be synovial sarcoma (194 cases) were included as negative controls. All cases were reviewed by MFCA, RT, AMF, and the cases that had no SS18 rearrangement were also reviewed by AH, CF and AN. The study complied with Brazilian and local ethical standards.
Representative paraffin-embedded material and haematoxylin and eosin-stained slides were reviewed. Mitotic figures were counted in 10 consecutives high-power fields (1 HPF=0.238 mm2) in highly proliferative ‘hot spot’ areas. Clinical data on selected cases were collected from histopathology reports.
Classification of Synovial Sarcoma
The diagnosis of synovial sarcoma was based on the morphological features and immunohistochemistry as defined previously.4, 38 All cases were classified as either monophasic or biphasic subtypes. The presence of well-defined glandular structure was required for the diagnosis of the latter, whereas monophasic synovial sarcoma was composed of spindle cells often with epithelioid areas. Poorly differentiated areas were defined by the presence of previously described features in five of more low-power fields: (1) cellular spindle and fascicular architecture closely simulating high-grade fibrosarcoma and malignant peripheral nerve sheath tumor with areas of extensive necrosis, perivascular tumor preservation and high-mitotic activity; (2) cellular areas composed of round cells with features intermediate between the spindle and epithelioid cells, sometimes with rhabdoid features; and (3) the small cell variant that closely resembles PNET/Ewing sarcoma.42
Immunohistochemistry
Immunohistochemistry was performed using the Ventana NexES Autostainer (Ventana Medical Systems, Strasbourg, France) using antibodies to α-smooth muscle actin (1A4, 1:25 dilution, Dako, High Wycombe, Hertfordshire, UK), desmin (DE-R-11, 1:10 dilution, 10 min protease pretreatment, Dako), S100 protein (polyclonal, 1:1600 dilution, 6 min protease pretreatment, Dako), EMA (E-29, 1:5 dilution, 2 min pressure cooker pretreatment, Dako), AE1/AE3 (dilution 1:50, 12 min protease pretreatment, Dako), MNF116 (dilution 1:100, 14 min protease pretreatment, Dako), CD 99 (clone 12E7, dilution 1:50, 2 min pressure pretreatment, Dako) and bcl-2 (clone M0887, dilution 1:10, 2 min pressure cooker pretreatment, Dako). Diaminobenzidine was used as a chromogen in all reactions. Positive and negative controls were included in each immunohistochemistry run.
Tissue Microarray
A tissue microarray was constructed using a manual tissue arrayer (Beecher Instruments Inc, Sun Prarie, WI, USA). Two different areas from each case were marked in the paraffin blocks and two 0.6 mm cores were taken for the array from all the cases described above for which material was available (113 cases). One hundred and seventeen cores including salivary gland, kidney, placenta and liver were used as controls and orientation markers.
RNA Extraction and Reverse Transcription
Two 10 μm or 5–10 10 μm sections were cut from resection or biopsy specimen blocks, respectively, and placed into Eppendorf tubes. RNA was extracted from paraffin-embedded samples using Optimum® FFPE RNA Isolation Kit (Ambion Europe, Huntington, Cambridgeshire, UK). Between 1 and 3 μl of the resulting RNA samples were reverse transcribed using Superscript III First-Strand Synthesis kit (Invitrogen, Inchinnan, Paisley, Scotland, UK) using gene-specific primers (all primers and product sizes shown in Table 1). A G6PD (glucose-6-phosphate dehydrogenase) anti-sense primer was included in the reverse transcription step.
Quantitative Polymerase Chain Reaction
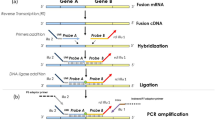
Quantitative polymerase chain reaction (qPCR) amplification was performed using the iCycler iQ detection system (Bio-Rad Laboratories, Hempstead, Hertfordshire, UK) on 1 μl aliquots of cDNA samples using specific primer sets (SYT-B-RT and SSX1-2-B) and probes designed based on the known fusion genes and break points (see Figure 1 and Table 1). The reaction mixtures were made up to 25 μl using 12.5 μl of iQ™ Supermix (Bio-Rad Laboratories, Hempstead, Hertfordshire, UK), 5 pmol of each primer and 1 pmol of FAM-labelled probe and ultra pure water. Thermal cycling parameters were as follows: 3 min at 95°C followed by 55 cycles of 15 s at 94°C and 1 min at 60°C Fluorescence was monitored during the 60°C annealing step.
Schematic diagram of the variants of the fusion sites described for SS18-SSX1 (a), SS18-SSX2 (b) and SS18-SSX4 (c). The usual fusions are present in the majority of cases (boxes). The fusions involve codon 410 on SS18 (ENSG00000141380*) and codon 111 on either SSX1 (ENSG00000126752*) or SSX2 (ENSG00000157950*). SS18 and SSX1/2/4 sequences adjacent to the break point are shown above each diagram and the exons and codons involved below. The positions of inserted sequences, not derived from these genes, are indicated by grey triangles. *Gene structure, exon and codon positions from Ensembl v39. http://www.ensembl.org/Homo_sapiens.
Restriction Enzyme Digestion for Determining Subtype of SS18-SSX Transcript Generated by qRT-PCR
Three microlitres of PCR product was mixed with 0.5 μl of restriction enzyme XmnI (New England Biolabs, Hitchin, Hertfordshire, UK), 1 μl of buffer (New England Biolabs) and made up to a final volume of 10 μl with ultra pure water and digested for 2 h at 37°C. The products were run through a 10% polyacrylamide gel revealing 56 and 39 bp products for a digested SS18-SSX2 transcript, and an undigested 95 bp product for the SS18-SSX1 fusion.30
Conventional Polymerase Chain Reaction
PCR amplification was performed on duplicate samples of 1 μl aliquots of cDNA using specific primer sets designed based on the known fusion genes and break points (SSA, SSX1, SSX2 and SSX4) (Figure 1) (Table 1).49 Reactions were made up in 25 μl using 1X buffer II (Applied Biosystems, Warrington, Cheshire, UK), 200 μM of each dNTP, 5 pmol of each primer, 1.5 mM MgCl2 and 1 U of Amplitaq Gold (Applied Biosystems). A touchdown protocol was used with cycling parameters as follows: 7 min at 95°C followed by 45 s at 94°C, 45 s at 66°C, 1 min 30 s at 72°C which was followed by reducing the annealing temperature by 1°C each cycle to 57°C (10 cycles), followed by 30 cycles at 56°C and finally 5 min at 72°C.
SS18-SSX1 and SS18-SSX2 translocations were sought using primer pairs SSA-SSX1 and SSA-SSX2, respectively (Table 1).49 Products were separated through an 8% polyacrylamide gel, stained with ethidium bromide and visualized under UV illumination using a BioRad Gel Doc 2000™ system (Hemel Hempstead, Hertfordshire, UK). Samples yielding PCR products of the predicted size (108 bp) in both reactions were scored as positive.
Controls for RT-PCR
A negative (no template) and positive control (SS18-SSX fusion gene confirmed by sequencing) were used for each experiment. The housekeeping gene, G6PD, was amplified using the same reaction conditions and cycling parameters for conventional RT-PCR, as described above. The PCR primers were designed to provide the template for generation of products of 86, 141 and 200 bp, from the housekeeping gene G6PD to control for RNA quality.
FISH
FISH was performed on the tissue microarray after optimization of the protocol using LSI SYT Dual Color Break-Apart Rearrangement Probe (Vysis, Abbott Laboratories Inc., Maidenhead, Berkshire, UK). One end of the probe was labeled with spectrum orange™ (telomeric, 5′ to SS18, 650 Kb) and the other with spectrum green™ (centromeric, 3′ to SS18, 1040 Kb). Sections were deparaffinized in xylene (3 × for 5 min) and dehydrated in ethanol (3 × for 3 min). Sections were pretreated using Paraffin Pretreatment Reagent Kit II (Vysis). This involved placing the slides in 50 μl of pretreatment solution at 80°C for 50 min, followed by 3 min in distilled water, 20 min in protease solution at 37°C washing in distilled water for 3 min and dehydrating for 1 min in increasing concentrations of alcohol (70, 85 and 100%) after which they were air-dried. Ten microlitres of probe mixture (1 μl probe, 2 μl dH2O and 7 μl hybridizing buffer) were applied to the slides, overlaid with a coverslip, which was sealed with rubber cement. Slides were denatured for 5 min at 73°C and hybridized for at least 16 h at 37°C in a ThermoBrite™ hybridizer (Iris, Westwood, MA, USA). Following hybridization, the slides were washed with 2 × saline sodium citrate (SCC)/0.3% NP40 at room temperature for 5 min, at 73°C for 2 min and again at room temperature for 1 min. After air-drying in the dark, slides were counterstained with 10 μl of a mixture of Vectashield® mounting medium (Vector Laboratories, Orton Southgate, Peterborough, UK) containing 5 μl of Vectashield® with 4′6′-diamino-2-phenylindole (DAPI) and 5 μl without DAPI. A coverslip was applied and sealed with nail varnish. The slides were analyzed under a fluorescent microscope (Olympus, Southall, Middlesex, UK) equipped with a mercury lamp, DAPI/FITC/Rhodamine triple filter and individual DAPI, FITC and rhodamine filters using AnalySIS software (Olympus). Fifty non-overlapping nuclei, which were clearly identified and contained unequivocal signals, were counted for each case. A probe was considered to be split when the orange and green signals were separated by a distance greater than the size of one hybridization signal (spectrum orange 650 kb): this cutoff was chosen because the distance between the non-split probes is 56 kb. When 20% or more single nuclei were found to harbor both a fusion and split (break-apart) signals, the cases were classified as positive and when less than 20% of cells had a fusion and split signals they were scored as negative.51 Scoring of the cases was performed without knowledge of the RT-PCR results.
Statistical Analysis
χ2 tests or Fisher exact tests were used, where applicable, to analyze differences between independent variants. Differences were considered significant if P<0.05. The software SPSS 10.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Results
Clinical and Histological Features
Synovial sarcoma was included in the differential diagnosis of 134 cases based on the morphology and immunohistochemistry: in 97 of these synovial sarcoma was considered the primary diagnosis and as a strong candidate in the remaining 37 cases. Patients with these tumors presented between the ages of 5 and 81, the mean age being 35 years, and the tumors occurred equally in both sexes (female/male ratio, 1:1.06). The lower extremity was the most common location (n=80), followed by upper extremity (n=25), thorax (n=17), head and neck (n=4), pelvis (n=2) and retroperitoneum (n=2). In four cases the location was unknown (Table 2). One hundred and three (77%) cases were classified as monophasic synovial sarcoma and 31 (23%) as biphasic synovial sarcoma. Poorly differentiated areas were identified in 44 (33%) cases (Figure 2, Table 3). When all of the 134 cases were analyzed, the number of mitotic figures per 10 HPF varied from 1 to 54 (mean 13.5), but when only cases with poorly differentiated areas were analyzed, the mitotic figures varied from 2 to 54 (mean 25.4). On further analysis of the 134 cases together, between 1 and 9 mitotic figures/10 HPF were found in 50% of the cases, 10-19/10 HPF in 22% and ≥20/10HPF in 28% of the cases. The areas with ≥20 mitotic figures/10 HPF were associated with the presence of poorly differentiated areas (P<0.001).
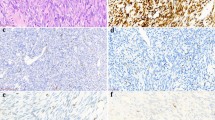
Photomicrographs of haematoxylin & eosin-stained sections and immunohistochemistry of synovial sarcomas. The variable microscopic appearance of haematoxylin & eosin-stained sections of synovial sarcoma (a–g) (original magnification × 200). Representative sections showing expression of bcl-2 in neoplastic cells (h), tumors cells which are negative for CD34 with vessels being decorated by the antibody (i), keratins (AE1/AE3) labeling both the epithelioid component (j) and isolated neoplastic spindle cells (k). EMA—membranous staining of tumors cells (l).
Tumor cells expressed cytokeratins (AE1/AE3 and/or MNF116) and/or EMA, at least focal, in 80 and 98% of cases, respectively. Bcl-2 and S100 protein expression was detected 98 and 22%, respectively, CD99 expression was observed in 56% of cases. The neoplastic cells were positive for CD34 in one case and were negative for the remaining 126 cases tested. No cases revealed desmin or α-smooth muscle actin immunoreactivity.
Detection of SS18-SSX Fusion Transcripts
Three of the 134 cases failed to give a result by qRT-PCR and RT-PCR using any of the primer sets: a PCR product for the housekeeping gene, G6PD, also failed to be generated in these cases (Table 1). One hundred and twenty-six of the 131 cases studied revealed SS18-SSX fusion transcripts by qRT-PCR (Figure 3). SS18-SSX fusion transcripts were not detected in the remaining five cases in which G6PD products as large as 200 bp were generated indicating amplifiable cDNA in the samples. Conventional RT-PCR generated products representing the usual variants of SS18-SSX1 or SS18-SSX2 in 119 of 131 cases. An additional case revealed a larger product which was reported previously as a novel SS18-SSX1 variant.26 Fusion genes failed to be detected in the remaining 11 cases by conventional RT-PCR using primers SSA, SSX1, SSX2 and SSX4 shown in Table 1. Five of these cases also failed to give a SS18-SSX fusion gene using primers SYT-B-RT and SSX1-2-B by qRT-PCR (Table 1). A SS18-SSX fusion gene was detected in three of the remaining six negative cases when employing qRT-PCR primers (see Table 1) in a conventional PCR assay. Restriction enzyme digestion found four and two of these cases to harbor a SS18-SSX1 and SS18-SSX2 fusion transcript, respectively.
Detection of SS18-SSX fusion transcripts by the RT-PCR. Thirty-four samples analyzed by qRT-PCR and represented by different lines: controls included (a). Detection of a positive SS18-SSX2 fusion transcript, by conventional RT-PCR in duplicate in an 8% polyacrylamide gel (b): lane L, molecular weight markers (50 bp); lane 1 and 5, negative control; lanes 2 and 6, positive controls SSX1 and SSX2 respectively; lanes 3 and 4, negative for SS18-SSX1; lanes 7 and 8, positive for SS18-SSX2.
SS18-SSX4, SS18LI-SSX135 transcripts and other variants of SS18-SSX1 and SS18-SSX2 (Figure 1 and Table 1) failed to be detected in the five negative cases (data not shown) using conventional PCR. A usual SS18-SSX fusion transcript was not detected in 194 cases where a diagnosis synovial sarcoma was largely excluded based on the morphology and immunohistochemistry, or by the detection of another diagnostic fusion transcript (Table 4). G6PD PCR products of at least 141 bp were generated in all of these cases by RT-PCR.
FISH
Of the 113 cases of synovial sarcoma, as defined by morphology and immunohistochemistry, included in the tissue microarray, 12 (11%) could not be analyzed owing to core loss (n=4) or absent/weak fluorescent signals (n=8). Of the 101 remaining cases, 87 (86%) showed SS18 rearrangement, and in these the percentage of cells with intact nuclei containing both a fused and break-apart signal varied from 21 to 74% (average 50%) (Figure 4). Fourteen cases failed to show SS18 rearrangement: five of these were also negative by conventional and qRT-PCR and nine showed a SS18-SSX fusion transcript by conventional RT-PCR and qRT-PCR (n=8) and qRT-PCR only (n=1). Analysis of four of these nine cases revealed that at least 70% of nuclei contained one fusion signal and a single spectrum orange signal, although a fusion and break-apart signals were identified in occasional cells (2, 4, 6 and 8%) (Figure 4). In the remaining five cases, which were scored as negative, a break-apart signal was detected in 2–4 % of the cells in three cases and no break-apart signal was detected in two cases. No cases were found to show between 10 and 20% of the cells with break-apart signals.
Schematic diagram of chromosome 18 showing the position of the SS18 gene and fluorescent probes (right side panel). Photomicrograph of representative FISH results (left side panel) and respective schematic diagrams (middle panel): two fused red and orange signals demonstrating no SS18 rearrangement (a); a break-apart signal revealing SS18 rearrangement (b); loss of the spectrum green signal (c), and multiple copies involving the SS18 region (d).
Fifteen of the 101 cases analyzed by FISH showed multiple copies of the SS18 gene (Figure 4). The intact nuclei of 14 of these cases showed more than 20% of break-apart signals. Case 15 was classified as negative on account of only 2% of cells showing unequivocal break-apart signals. However, this case was positive by both conventional and qRT-PCR.
One of the three cases, which could not be analyzed by RT-PCR, owing to no amplifiable cDNA, was positive by FISH. One was not tested by FISH owing to unavailable material and the remaining case was tested but no signal was detected.
Correlation of SS18-SSX Fusion Genes and Morphology
Collation of all the data generated from conventional and qRT-PCR revealed that 74 and 52 of the cases were found to harbor SS18-SSX1 and SS18-SSX2 fusion genes, respectively. The relationship between histological subtypes (mono or biphasic) and fusion transcripts is shown in Table 3, and was not found to be statistically significant (P=0.9). The presence of poorly differentiated areas was identified in both biphasic (n=5, 16%) and monophasic (n=39, 38%) neoplasms and did not correlate with fusion type (P=0.2).
Of the 31 biphasic synovial sarcoma, 30 showed areas of unequivocal glandular formation. One case which showed extensive squamous differentiation harbored a SS18-SSX2 fusion type.
SS18-SSX-Negative Cases
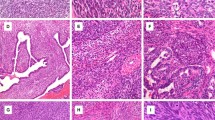
Table 5 and Figure 5 provide details of the 5 SS18-SSX fusion gene negative cases. Of the three pleural cases, one exhibiting a biphasic and two a monophasic pattern, the diagnosis of synovial sarcoma was favored based on morphology in cases one and two initially. However, following clinical and radiological input into case 1, it was considered that ‘on the balance of probabilities’ the tumor was a biphasic mesothelioma and not a synovial sarcoma. This decision was made in view of the unilateral disease, the history of asbestos exposure and an immunophenotype that overlapped between synovial sarcoma and mesothelioma, despite the atypical features for synovial sarcoma, which included very extensive expression of cytokeratins in the spindle cell areas. In case 2, the tumor was a more poorly differentiated neoplasm diffusely positive for bcl-2 protein and CD99, but negative for cytokeratins and other markers. EWS gene rearrangement was not detected by either FISH or RT-PCR (data not shown) and this tumor was eventually classified as a sarcoma, not otherwise specified, despite a history of asbestos exposure. The third pleural tumor was thought most likely to represent a solitary fibrous tumor based on the haematoxylin and eosin sections and radiological appearances but as the tumor was negative for CD34 and CD99 (Table 5), it was reassuring for a SS18 rearrangement not to be detected. In the remaining two cases, one was an undifferentiated small cell neoplasm situated in the pelvis: it was negative for SS18 and EWS rearrangements and was classified as an undifferentiated sarcoma, not otherwise specified. Synovial sarcoma remained the favored diagnosis for the fifth case, despite histological review and the failure to detect a SS18-SSX rearrangement. However, in the absence of a SS18-SSX rearrangement, a malignant peripheral nerve sheath tumor could not be excluded in cases 4 and 5.
Photomicrographs of haematoxylin & eosin-stained sections (original magnification × 200) showing the histological appearance of 5 SS18-SSX negative cases (cases 1 to 5—Table 5).
Discussion
This study of 134 tumors, in which synovial sarcoma was the favored diagnosis or was considered high in the differential diagnosis under transmitted light microscopy, shows that qRT-PCR, conventional RT-PCR and FISH are all valuable means of detecting SS18-SSX gene rearrangements—the hallmark of synovial sarcoma. Only three of 134 cases failed to render cDNA of sufficient quality for analysis by PCR showing that formal-fixed paraffin-embedded material is almost always suitable for use in these assays in a diagnostic laboratory. Nevertheless, in our hands, qRT-PCR is the most sensitive technology for detecting the genetic abnormality involving SS18, identifying 126 of 131 (96%) cases compared with 120 of 131 (92%) cases using conventional RT-PCR. FISH was the least sensitive approach for detecting these transcripts with the study revealing that only 87 of 101 (86%) cases gave a positive result. However, in view of one case giving a positive result by FISH, which could not be analyzed by RT-PCR, owing to failure of amplification of cDNA, FISH is considered to complement RT-PCR. When the three cases, which could not be analyzed owing to insufficient cDNA quality, were included, the sensitivity would be 94% using qRT-PCR and 89.5% using conventional RT-PCR.
In addition to FISH in this study being less sensitive than RT-PCR for detecting SS18 gene rearrangements, problems of interpreting the FISH were also encountered. These difficulties are largely three-fold: the first, not previously reported, is the detection of one fusion signal (green and orange) and a single spectrum orange signal, in more than 70% of the neoplastic cells, in other words, the second spectrum green label was not observed. This observation was made in four cases, in which the SS18-SSX fusion gene was detected by RT-PCR, and has been found in a further two cases since the study was closed. The SSX-SS18 transcript, unlike its reciprocal SS18-SSX fusion gene, has been shown to be lost in as many as two-thirds of cases of synovial sarcoma, and this loss has been associated with progression of disease.27 We, therefore, speculate that our observation is caused by deletion of the SSX-SS18 fusion gene. Our interpretation is also based on the fact that the SS18-SSX fusion is composed mainly of the SS18 gene, to which the spectrum orange label is attached via the 5′-end, and spectrum green labels the SSX-SS18 fusion. Nevertheless, further studies are required to support this theory.
The second difficulty encountered when interpreting FISH for the diagnosis of synovial sarcoma is the presence of multiple signals in some cases: this finding represents either polyploidy or multiple copies of a region of variable length of chromosome 18 in the region of the SS18 gene. This has been reported previously in synovial sarcoma, with one of the most common gains involving 18q,27 and also in PNET/Ewing sarcoma in chromosome 22.52 On occasions the signals are so numerous that the result is difficult to interpret although in 14 of our 15 cases unequivocal break-apart signals were also identified allowing synovial sarcoma to be diagnosed. In all of these cases, interpretation of the FISH result was supported by RT-PCR.
The final problem experienced using FISH is determining (a) how many cells should be counted and (b) what percentage of these cells need to be scored as positive for break-apart signals before a tumor is classed as harboring a rearrangement. In this study, we employed the standards of Lu et al,51 but we did not encounter cases where the cells with break-apart signals represented between 8 and 21% of the 50 cells counted. In borderline cases, it is useful to have supportive information from RT-PCR analysis but most importantly any result must be interpreted in the light of the morphology and immunohistochemistry. For these reasons, we believe that there are great benefits for the histopathologists reporting such cases to have experience in the interpretation of FISH and have direct access to this technology.
Our results demonstrate that the primers and probes used for RT-PCR in this study are specific for the various subtypes and variants of the reported SS18-SSX fusion genes. Not only were SS18-SSX fusion transcripts not detected in 194 other tumors by RT-PCR (109 tumors in which other fusion genes were detected and 85 other soft tissue and bone tumors with no fusion detected) but there was also complete concordance of positive results when using primers for conventional and qRT-PCR. In this study and in all other series except one,53 the type of transcripts (SS18-SSX1 or SS18-SSX2) was mutually exclusive and persisted in recurrent and metastatic disease (data not shown). It is difficult to explain the results of Yang et al as their study was in-depth and appears robust. Nevertheless, it is important to highlight that SSX1 and SSX2 share at least 80% homology,21 and that primer design is therefore crucial if the two genes are to be distinguished by PCR.
The fact that six cases in this study were found to harbor the SS18-SSX fusion transcript using qRT-PCR, but not conventional RT-PCR suggests that the former technology is more sensitive. However, this needs to be interpreted critically as the data show that at least some of this apparent greater sensitivity is based on the PCR primer design and possibly the size of the product generated rather than the technology employed.
This study reveals that biphasic synovial sarcoma not uncommonly harbors the SS18-SSX2 fusion, a finding that contrasts results of previous series in which almost all biphasic cases were shown to harbor a SS18-SSX1 fusion type.21 However, having performed a meta-analysis of published data we find that SS18-SSX2 occurs significantly more frequently in monophasic than in biphasic synovial sarcoma (P<0.001): this meta-analysis included a total of 1033 cases, of which 314 were biphasic synovial sarcoma and only 42 of these harbored the SS18-SSX2 transcript.5, 20, 22, 27, 30, 37, 39, 46, 47, 49, 50, 51, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65 The low frequency of both the SS18-SSX2 fusion type and the biphasic subtype of synovial sarcoma best explain the discrepant results observed in various studies. In our study, we also failed to find a significant association between poorly differentiated areas of synovial sarcoma and SS18 gene rearrangement, thereby supporting the work by Guillou et al.39
This study highlights the difficulty in distinguishing synovial sarcoma from mesothelioma, an important issue as the thorax is now recognized as a not uncommon site in which synovial sarcoma occur5, 43 and the correct classification of tumors not only alters clinical management but in the case of mesothelioma, the patient may also be eligible for financial compensation if there has been exposure to asbestos. We found that on microscopic assessment alone it was not possible to distinguish definitively between these two tumor types and detection of a SS18 rearrangement would have been invaluable in confidently refuting the diagnosis of mesothelioma. Of the five SS18-SSX negative neoplasms in this study, three were pleural lesions, one exhibiting a biphasic and two a monophasic pattern. A diagnosis of synovial sarcoma was strongly favored in two of these three cases when the histopathology was analyzed in the absence of further clinical information. A diagnosis of a solitary fibrous tumor was favored in the third case and molecular genetic analysis was performed for exclusion purposes. However, when the histopathology was reviewed in the light of all the information from the multidisciplinary team, which included physicians, radiologists and pathologists, and the negative results from the molecular analysis were taken into account, all three cases were considered unlikely to represent synovial sarcoma. The value of a multidisciplinary approach is emphasized further as one pleural case was eventually considered to be sarcoma ‘not otherwise specified’ rather than mesothelioma despite exposure to asbestos.
Of the two non-pleural-based SS18-SSX negative tumors, one, situated in the pelvis, was classified as a small cell undifferentiated neoplasm, not otherwise specified, and the second occurred in the lower limb. Despite histological review of the latter and the failure to detect SS18-SSX rearrangements the diagnosis of synovial sarcoma was still favored. Nevertheless, as there is a well-recognized overlap both morphologically and at a gene expression level between synovial sarcoma and malignant peripheral nerve sheath tumor66, 67 the latter was seriously considered as the diagnosis in both of these cases. However, as neither of these patients had stigmata of neurofibromatosis, and the tumors did not obviously arising from a large nerve this diagnosis was not possible to make with confidence.
If aberrant genetic events are to be employed as diagnostic tests on which clinical management is to be based, knowledge of the sensitivity and specificity of any method being employed is crucial. The majority of published evidence, including ours, argues that detection of a SS18 gene rearrangement is specific for the diagnosis of synovial sarcoma,48, 68 although there is some debate over the interpretation of finding a SS18 fusion gene in tumors with morphological features of malignant peripheral nerve sheath tumors.69, 70, 71 In contrast, proving 100% sensitivity is more difficult. Indeed, several groups, including ours, demonstrate it is possible to attain at least 96% sensitivity for the detection of a SS18 gene rearrangement but none claim 100%.21, 39, 49, 50, 59 For this to be achieved in our study, it would be necessary to demonstrate, beyond all doubt, the nature of the five cases which had morphological features of synovial sarcoma but in which a SS18 gene rearrangement was not detected. Unfortunately, a truly robust test for diagnosing mesothelioma, malignant peripheral nerve sheath tumor and solitary fibrous tumor, the favored diagnoses for our five cases when a SS18 rearrangement was not detected, does not exist, and therefore claiming 100% sensitivity is likely to result in development of dogma which tend not to be questioned—a dangerous mind-set for pathologists. Although we have screened for all the known possible variants, the possibility of the existence of a novel t(X;18) variant remains, alternatively the failure to detect a SS18 rearrangement might be explained by almost complete loss of the X chromosome something which has been described in advanced tumors.54 Hence, we report these rare cases descriptively, and on the basis of clinical and radiological information.
Both FISH and RT-PCR have advantages and disadvantages, but this study shows that they complement each other. FISH is often favored by pathologists as they are more familiar with analyzing cells using a microscope than reading gels. However, unless specific probes for fusion variants are available, FISH is less informative than RT-PCR. This begs the question ‘what information is required from molecular diagnostic investigations?’ The answer will vary from tumor to tumor, and may change over time: for example, if it is demonstrated that identification of a fusion variant for a particular tumor does not alter treatment and provides no information about clinical outcome, identification of specific variants may be considered irrelevant. Therefore, continued critical assessment of why specific investigations are undertaken is crucial if pathologists are to use available resources efficiently.
References
Russell WO, Cohen J, Edmonson JH, et al. Staging system for soft tissue sarcoma. Semin Oncol 1981;8:156–159.
Cadman NL, Soule EH, Kelly PJ . Synovial sarcoma. Cancer 1965;18:613–627.
Cagle LA, Mirra JM, Storm FK, et al. Histologic features relating to prognosis in synovial sarcoma. Cancer 1987;59:1810–1814.
Weiss SW, Goldblum JR . Synovial sarcoma. In: Weiss SW, Goldblum JR (eds). Enzinger and Weiss's Soft Tissue Tumors. Mosby: London, 2001, pp 309–346.
Begueret H, Galateau-Salle F, Guillou L, et al. Primary intrathoracic synovial sarcoma: a clinicopathologic study of 40 t(X;18)-positive cases from the French Sarcoma Group and the Mesopath Group. Am J Surg Pathol 2005;29:339–346.
Fisher C, Folpe AL, Hashimoto H, et al. Intra-abdominal synovial sarcoma: a clinicopathological study. Histopathology 2004;45:245–253.
Hisaoka M, Hashimoto H, Iwahana H, et al. Primary synovial sarcoma of the lung: report of two cases confirmed by molecular detection of SYT-SSX fusion gene transcripts. Histopathology 1999;34:205–210.
Holtz F, Magielski JE . Synovial sarcomas of the tongue base. Arch Otolaryngol 1985;111:271–272.
Iyengar V, Lineberger AS, Kerman S, et al. Synovial sarcoma of the heart. Correlation with cytogenetic findings. Arch Pathol Lab Med 1995;119:1080–1082.
Jun SY, Kang GH, Park SH, et al. Synovial sarcoma of the kidney with rhabdoid features: report of three cases. Am J Surg Pathol 2004;28:634–637.
Lenoir P, Ramet J, Goossens A, et al. Retropharyngeal synovial sarcoma in an infant: report of a case and its response to chemotherapy; review of the literature. Pediatr Hematol Oncol 1991;8:45–52.
Ng SB, Ahmed Q, Tien SL, et al. Primary pleural synovial sarcoma. A case report and review of the literature. Arch Pathol Lab Med 2003;127:85–90.
Nicholson A, Rigby M, Lincoln C, et al. Synovial sarcoma of the heart. Histopathology 1997;30:349–352.
O'Donnell P, Diss T, Whelan J, et al. Synovial sarcoma with radiological appearances of primitive neuroectodermal tumour/Ewing sarcoma: differentiation by molecular genetic studies. Skeletal Radiol 2006;35:233–239.
Okamato S, Hisaoka M, Daa T, et al. Primary pulmonary synovial sarcoma: a clinicopathologic, immunohistochemical, and molecular study of 11 cases. Human Pathol 2004;35:850–856.
Limon J, Mrozek K, Nedoszytko B, et al. Cytogenetic findings in two synovial sarcomas. Cancer Genet Cytogenet 1989;38:215–222.
Turc-Carel C, Dal Cin P, Limon J, et al. Involvement of chromosome X in primary cytogenetic change in human neoplasia: nonrandom translocation in synovial sarcoma. Proc Natl Acad Sci USA 1987;84:1981–1985.
Turc-Carel C, Dal CP, Limon J, et al. Translocation X; 18 in synovial sarcoma. Cancer Genet Cytogenet 1986;23:93.
Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994;7:502–508.
Crew AJ, Clark J, Fisher C, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995;14:2333–2340.
Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002;62:135–140.
Nilsson G, Skytting B, Xie Y, et al. The SYT-SSX1 variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res 1999;59:3180–3184.
Safar A, Wickert R, Nelson M, et al. Characterization of a variant SYT-SSX1 synovial sarcoma fusion transcript. Diagn Mol Pathol 1998;7:283–287.
Sanders ME, van de Rijn M, Barr FG . Detection of a variant SYT-SSX1 fusion in a case of predominantly epithelioid synovial sarcoma. Mol Diagn 1999;4:65–70.
O'Sullivan MJ, Humphrey PA, Dehner LP, et al. t(X;18) reverse transcriptase-polymerase chain reaction demonstrating a variant transcript. Mol Diagn 2002;4:178–180.
Amary MFC, Diss T, Flanagan AM . Molecular characterization of a novel variant of a SYT-SSX1 fusion transcript in synovial sarcoma. Histopathology 2006, in press.
Panagopoulos I, Mertens F, Isaksson M, et al. Clinical impact of molecular and cytogenetic findings in synovial sarcoma. Genes Chromosomes Cancer 2001;31:362–372.
Fligman I, Lonardo F, Jhanwar SC, et al. Molecular diagnosis of synovial sarcoma and characterization of a variant SYT-SSX2 fusion transcript. Am J Pathol 1995;147:1592–1599.
Tornkvist M, Brodin B, Bartolazzi A, et al. A novel type of SYT/SSX fusion: methodological and biological implications. Mod Pathol 2001;15:679–685.
Gaffney R, Chakerian A, O'Connell JX, et al. Novel fluorescent ligase detection reaction and flow cytometric analysis of SYT-SSX fusions in synovial sarcoma. Mol Diagn 2003;5:127–135.
Skytting B, Nilsson G, Brodin B, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 1999;91:974–975.
Mancuso T, Mezzelani A, Riva C, et al. Analysis of SYT-SSX fusion transcripts and bcl-2 expression and phosphorylation status in synovial sarcoma. Lab Invest 2000;80:805–813.
Brodin B, Haslam K, Yang K, et al. Cloning and characterization of spliced fusion transcript variants of synovial sarcoma: SYT/SSX4, SYT/SSX4v, and SYT/SSX2v. Possible regulatory role of the fusion gene product in wild type SYT expression. Gene 2001;268:173–182.
Agus V, Tamborini E, Mezzelani A, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Instit 2001;93:1347–1349.
Storlazzi CT, Mertens F, Mandahl N, et al. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer 2003;37:195–200.
Sonobe H, Takeuchi T, Liag SB, et al. A new human synovial sarcoma cell line, HS-SY-3, with a truncated form of hybrid SYT/SSX1 gene. Int J Cancer 1999;82:459–464.
Wei Y, Wang J, Zhu X, et al. Detection of SYT-SSX fusion transcripts in paraffin-embedded tissues of synovial sarcoma by reverse transcrition-polymerase chain reaction. Chin Med J (Engl) 2002;115:1043–1047.
Fisher C . Synovial sarcoma. Ann Diagn Pathol 1998;2:401–402.
Guillou L, Benhattar J, Bonichon F, et al. Histologic grade, but not SYT-SSX fusion type, is an important prognostic factor in patients with synovial sarcoma: a multicenter, retrospective analysis. J Clin Oncol 2004;22:4040–4050.
Ladanyi M, Bridge JA . Contribution of molecular genetic data to the classification of sarcomas. Human Pathol 2000;31:532–538.
Skytting B . A Scandanavian sarcoma group project. Acta Orthop Scand Suppl 2000;291:1–28.
Meis Kindblom JM, Stenman G, Kindblom LG . Differential diagnosis of small round cell tumors. Semin Diagn Pathol 1996;13:213–241.
Folpe AL, Schmidt RA, Chapman D, et al. Poorly differentiated synovial sarcoma: immunohistochemical distinction from primitive neuroectodermal tumors and high-grade malignant peripheral nerve sheath tumors. Am J Surg Pathol 1998;22:673–682.
de Silva MV, McMahon AD, Paterson L, et al. Identification of poorly differentiated synovial sarcoma: a comparison of clinicopathological and cytogenetic features with those of typical synovial sarcoma. Histopathology 2003;43:220–230.
Coindre JM, Pelmus M, Hostein I, et al. Should molecular testing be required for diagnosing synovial sarcoma? A prospective study of 204 cases. Cancer 2003;98:2700–2707.
Thorson JA, Weigelin HC, Ruiz RE, et al. Identification of SYT-SSX transcripts from synovial sarcomas using RT-multiplex PCR and capillary electrophoresis. Mod Pathol 2006;19:641–647.
Tvrdik D, Povysil C, Svatosova J, et al. Molecular diagnosis of synovial sarcoma: RT-PCR detection of SYT-SSX1/2 fusion transcripts in paraffin-embedded tissue. Med Sci Monit 2005;11:MT1–MT7.
dos Santos NR, De Bruijn DR, Van Kessel AG . Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 2001;30:1–14.
Naito N, Kawai A, Ouchida M, et al. A reverse transcriptase-polymerase chain reaction assay in the diagnosis of soft tissue sarcomas. Cancer 2000;89:1992–1998.
Bijwaard KE, Fetsch JF, Przygodzki R, et al. Detection of SYT-SSX fusion transcripts in archival synovial sarcomas by real-time reverse transcriptase-polymerase chain reaction. Mol Diagn 2002;4:59–64.
Lu YJ, Birdsall S, Summersgill B, et al. Dual colour fluorescence in situ hybridization to paraffin-embedded samples to deduce the presence of the der(X)t(X;18)(p11.2;q11.2) and involvement of either the SSX1 or SSX2 gene: a diagnostic and prognostic aid for synovial sarcoma. J Pathol 1999;187:490–496.
Patel RM, Downs-Kelly E, Weiss SW, et al. Dual-color, break-apart fluorescence in situ hybridization for EWS gene rearrangement distinguishes clear cell sarcoma of soft tissue from malignant melanoma. Mod Pathol 2005;18:1585–1590.
Yang K, Lui WO, Xie Y, et al. Co-existence of SYT-SSX1 and SYT-SSX2 fusions in synovial sarcomas. Oncogene 2002;21:4181–4190.
Guillou L, Coindre JM, Gallagher G, et al. Detection of the synovial sarcoma translocation t(X;18) (SYT;SSX) in paraffin-embedded tissues using reverse transcriptase-polymerase chain reaction: a reliable and powerful diagnostic tool for pathologists. A molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Human Pathol 2001;32:105–112.
Antonescu CR, Kawai A, Leung DH, et al. Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol 2000;9:1–8.
Hill DA, O'Sullivan MJ, Zhu X, et al. Practical application of molecular genetic testing as an aid to the surgical pathologic diagnosis of sarcomas: a prospective study. Am J Surg Pathol 2002;26:965–977.
Hiraga H, Nojima T, Abe S, et al. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998;7:102–110.
Kawai A, Woodruff J, Healey JH, et al. Gene fusion as a determinant of morphology and prognosis in synovial sarcoma. New Engl J Med 1998;338:153–160.
Lasota J, Jasinski M, Biec-Rychter M, et al. Detection of the SYT-SSX fusion transcripts in formaldehyde-fixed, paraffin-embedded tissue: a reverse transcription polymerase chain reaction amplification assay useful in the diagnosis of synovial sarcoma. Mod Pathol 1998;11:626–633.
Mezzelani A, Mariani L, Tamborini E, et al. SYT-SSX fusion genes and prognosis in synovial sarcoma. Br J Cancer 2001;85:1535–1539.
Nikiforova MN, Groen P, Mutema G, et al. Detection of SYT-SSX rearrangements in synovial sarcomas by real-time one-step RT-PCR. Pediatr Dev Pathol 2005;8:162–167.
Shipley J, Crew J, Birdsall S, et al. Interphase fluorescence in situ hybridization and reverse transcription polymerase chain reaction as a diagnostic aid for synovial sarcoma. Am J Pathol 1996;148:559–567.
Tamborini E, Agus V, Mezzelani A, et al. Identification of a novel spliced variant of the SYT gene expressed in normal tissues and in synovial sarcoma. Br J Cancer 2001;84:1087–1094.
Tsuji S, Hisaoka M, Morimitsu Y, et al. Detection of SYT-SSX fusion transcripts in synovial sarcoma by reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Am J Pathol 1998;153:1807–1812.
Willeke F, Mechtersheimer G, Schwarzbach M, et al. Detection of SYT-SSX1/2 fusion transcripts by reverse transcriptase-polymerase chain reaction (RT-PCR) is a valuable diagnostic tool in synovial sarcoma. Eur J Cancer 1998;34:2087–2093.
Henderson SR, Guiliano D, Presneau N, et al. A molecular map of mesenchymal tumors. Genome Biol 2005;6:R76.
Nagayama S, Katagiri T, Tsunoda T, et al. Genome-wide analysis of gene expression in synovial sarcomas using a cDNA microarray. Cancer Res 2002;62:5859–5866.
van de Rijn M, Barr FG, Collins FG, et al. Absence of SYT-SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Pathol 1999;112:43–49.
O'Sullivan MJ, Kyriakos M, Zhu X, et al. Malignant peripheral nerve sheath tumors with t(X;18). A pathologic and molecular genetic study. Mod Pathol 2000;13:1336–1346.
Vang R, Biddle D, Harrison WR, et al. Malignant peripheral nerve sheath tumor with a t(X;18). Arch Pathol Lab Med 2000;124:864–867.
Tamborini E, Agus V, Perrone F, et al. Lack of SYT-SSX fusion transcripts in malignant peripheral nerve sheath tumors on RT-PCR analysis of 34 archival cases. Lab Invest 2002;82:609–618.
Acknowledgements
This work could not have been done without the work and co-operation of all the clinicians and radiologists in the London Bone and Soft Tissue Sarcoma Service. Thank you to those who sent cases for molecular analysis as Professor M Novelli at UCH, Dr AG Gibson at Kent & Canterbury Hospital NHS Trust, Dr Richard Ball at Norfolk and Norwich University Hospital NHS Trust, Tom Treasure at Guy's Hospital, and Professor NA Athanasou at Nuffield Orthopaedic Centre, Oxford.
Grant support: R&D funding from The Royal National Orthopaedic Hospital, Stanmore, UK.
MFC Amary* Research fellow supported by Coordination for the Improvement of Higher Education Personnel, CAPES—Brazil. PhD Student University of Sao Paulo, SP, Brazil.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amary, M., Berisha, F., Bernardi, F. et al. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol 20, 482–496 (2007). https://doi.org/10.1038/modpathol.3800761
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800761
Keywords
This article is cited by
-
Novel characteristics for immunophenotype, FISH pattern and molecular cytogenetics in synovial sarcoma
Scientific Reports (2023)
-
Predominantly epithelial-type synovial sarcoma with overwhelming neuroendocrine differentiation: a potential diagnostic pitfall
Diagnostic Pathology (2022)
-
Identification of novel SSX1 fusions in synovial sarcoma
Modern Pathology (2022)
-
EWSR1-SMAD3 fibroblastic tumour of bone: expanding the clinical spectrum
Skeletal Radiology (2021)
-
Correlating SS18-SSX immunohistochemistry (IHC) with SS18 fluorescent in situ hybridization (FISH) in synovial sarcomas: a study of 36 cases
Virchows Archiv (2021)