Abstract
Clear cell ependymoma is a rare and diagnostically challenging subtype of ependymoma, whose genetic features are essentially unknown. We studied 13 clear cell ependymomas (five cases WHO grade II, eight cases WHO grade III) by comparative genomic hybridization (CGH). Chromosomal imbalances were found in 12/13 cases. The most common aberrations overall were +1q (38%), −9 (77%), −3 (31%), and −22q (23%). Clear cell ependymomas of WHO grade II were characterized by −9 (40%), whereas WHO grade III cases mainly showed +1q (63%), and +13q (25%), as well as −9 (100%), −3 (38%), and −22q (25%). In contrast to other ependymal tumors, clear cell ependymomas of WHO grade II showed fewer imbalances than WHO grade III samples (1.4 vs 3.5 per case). Although some of the implicated chromosomes have previously been shown to be involved in other ependymoma variants, the striking frequency of +1q, −9, and −3 suggests that aberrations differ between clear cell and other types of ependymomas, in particular, for loss of chromosome 9 which can be regarded as the molecular hallmark of clear cell ependymomas.
Similar content being viewed by others
Main
Clear cell ependymomas were first reported by Kawano et al1 in 1983 as ‘ependymomas with oligodendroglioma-like cells’, and were later given the name ‘clear-cell ependymoma’ by the same authors.2 Although ependymomas are fairly frequent brain tumors, in particular, in the pediatric population where they are the third most common entity and account for approximately 10% of cases,3 the clear cell variety is exceedingly rare and diagnostically challenging.4, 5, 6, 7 Not surprisingly, only one study investigating the genetic properties of clear cell ependymomas has been undertaken by using fluorescence in situ hybridization (FISH) on 10 cases.5
As genetic analysis of clear cell ependymomas may provide biologically and diagnostically useful information, we carried out a comparative genomic hybridization (CGH) analysis of 13 cases.
Materials and methods
Patients and Tumors
Formalin-fixed and paraffin-embedded biopsy specimens of 13 primary clear cell ependymomas (nine males, four females; mean age: 12 years, range: 4–30 years) were investigated (Table 1). These consisted of five cases of WHO grade II (four males, one female; mean age: 14 years, range: 7–30 years), and eight of WHO grade III (five males, three females; mean age: 11 years, range: 4–25 years). Of these, 12 tumors were located supratentorially (92%). All patients underwent primary surgery, resulting in gross total resection in eight cases, and subtotal resection in five cases. Eleven patients underwent a combination of postoperative radiation therapy plus chemotherapy, while two patients did not receive any adjuvant therapy (Table 1). Clinical follow-up data were available for all 13 patients.
Histologic criteria for classifying a tumor as clear cell ependymoma were those employed by previous investigators.2, 4, 5, 7, 8 Briefly, clear cell ependymomas were demarcated and architecturally solid lesions characterized by sheets of uniform tumor cells with perinuclear halo and centrally located rounded nuclei. Focally, perivascular pseudorosettes were encountered. Tumors showed varying amounts of clear cell areas, and only specimens with at least 50% (and up to 100%) clear cell pattern per sample were included. Some tumors showed increased cellularity, palisading necroses, endothelial hyperplasia, and brisk mitotic activity, features indicative of anaplastic subtype.
Routine hematoxylin–eosin and elastica-van Gieson staining as well as immunohistochemistry using an avidin–biotin complex (ABC) technique and antibodies against glial fibrillary acid protein (GFAP), epithelial membrane antigen (EMA), vimentin, and NeuN were performed. Independent diagnoses were made by all three neuropathologists (CHR, AK, WP). Statistical analysis was performed using Student's t-test and χ2 test.
CGH Analysis
DNA was isolated by phenol–chloroform extraction according to standard protocols. CGH analysis was performed as described previously.9, 10 Briefly, tumor DNA was labelled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and reference DNA from a healthy male donor with digoxigenin-11-dUTP (Boehringer Mannheim) in a standard nick translation reaction. The DNAse concentration in the labelling reaction was adjusted to reveal an average fragment size of 200–500 base pairs. Labelled DNA fragments were purified from remaining nucleotides by column chromatography.
For CGH, 500 ng of tumor DNA, 300 ng of reference DNA and 30 μg of human Cot1 DNA (Gibco, Karlsruhe, Germany) were co-precipitated and redissolved in 10 μl of hybridization buffer. Denaturation of DNA was followed by a preannealing step of 45 min at 37°C. Target metaphase spreads which had been prepared following standard procedures, were denatured separately in 70% formamide/2 × SSC for 2 min at 72°C. Hybridization was allowed to proceed for 3–4 days. Post-hybridization washes were carried out at a stringency of 50% formamide/2 × SSC at 45°C and 0.1 × SSC at 60°C. Biotinylated and digoxigenated sequences were detected simultaneously, using avidin-FITC (Boehringer Mannheim, 1:200) and anti-digoxigenin-rhodamine (Boehringer Mannheim, 1:40). Slides were counterstained with DAPI and mounted in antifade solution (Vectashield, Vector Laboratories).
Microscopy and Digital Image Analysis
Separate digitized gray level images of DAPI, FITC and rhodamine fluorescence were taken with a CCD camera connected to a Leica DMRBE microscope. Image processing was carried out using the Applied Imaging Software. Average green-red ratios were calculated for each chromosome in at least 10 metaphases.
Chromosomal regions with CGH ratio profiles surpassing the 50% CGH thresholds (upper threshold 1.25, lower threshold 0.75) were defined as loci with copy number gains or losses. Over-representations were diagnosed as high-level gains or amplifications when the fluorescence intensity levels exceeded 1.5. As tumor specimens and normal DNA were not sex matched, X and Y chromosomes were excluded, as were centromeric and satellite regions of acrocentric chromosomes.
Results
Clear cell features ranged from focal to diffuse, although they generally comprised the majority of the tumor (Figure 1a). Immunohistochemically, all tumors were varyingly reactive for GFAP and vimentin, and showed occasional dot-like reaction for EMA, whereas membranous staining of the latter antibody was not seen (Figure 1b–d). No positivity was encountered for the neuronal marker NeuN.
Photomicrographs of a clear cell ependymoma showing sheets of uniform tumor cells with perinuclear halo, centrally located rounded nuclei, and occasional perivascular pseudorosettes (a), as well as immunohistochemical reactions for GFAP (b), vimentin (c), and dot-like staining for EMA (d). Original magnifications × 400 (a–c) and × 630 (d).
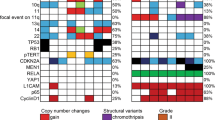
CGH among clear cell ependymomas revealed DNA copy number changes in 12/13 cases, with eight tumors showing at least one gain, and 12 cases with at least one loss (Table 1). The most common aberrations overall were +1q (38%), +4q, and +13q (15% each), as well as −9 (77%), −3 (31%), and −22q (23%). Clear cell ependymomas of WHO grade II were characterized by −9 (40%), whereas WHO grade III cases mainly showed +1q (63%), and +13q (25%), as well as −9 (100%), −3 (38%), and −22q (25%).
The highest number of DNA copy number changes was detected in a clear cell ependymoma WHO grade III (case 12: 7 imbalances), the lowest in a WHO grade II tumor (case 3: none). Clear cell ependymomas of WHO grade II showed fewer imbalances per case than WHO grade III samples (1.4 vs 3.5). Loss of chromosomal material was overall twice as frequent as gain (1.8 vs 0.9 per case), more so in WHO grade II cases (1.2 vs 0.2).
High-level gains only affected chromosome arm 1q and were exclusively encountered among clear cell ependymomas of WHO grade III (four out of eight cases).
Comparing the distribution and frequency of specific chromosomal imbalances between pediatric and adult cases, it was noticable that the three adult patients (cases 5, 12, and 13) showed some unusual chromosomal imbalances not matching the other cases, for example, gains of 5 and 13q as well as losses of 2, 6q, 10, 14q, and 17.
Clinical follow-up data were available for all 13 patients. At the time of reporting, one of five patients with WHO grade II tumors (20%, 44 months) and five of eight patients with WHO grade III tumors (63%, mean: 54 months, range: 34–88 months) had died of their disease (total 46%). The difference was not statistically significant (P=0.13). The number of aberrations overall as well as of gains and losses also bore no significance on survival.
Discussion
Clear cell ependymomas were first reported by Kawano et al1 in 1983 as ‘ependymomas with oligodendroglioma-like cells’, were later given the name ‘clear-cell ependymoma’,2 and were eventually included in the WHO classification of brain tumors.11 Clinically, they have a propensity for arising in the supratentorial compartment, which was found to be the case in eight of eight 8 and nine of 10 patients,5 a similar ratio to the 12/13 cases of our cohort. Furthermore, clear cell ependymomas seem to have an aggressive nature exemplified by their preponderance for recurrence,5 as encountered in 10 out of our 13 cases (Table 1).
Histologically, clear cell ependymomas are characterized by sheets of uniform tumor cells with perinuclear halo, centrally located rounded nuclei, and occasional perivascular pseudorosettes.2, 4, 5, 7, 8 However, there is no precise definition of how much clear cell area there has to be within a given tumor to warrant a diagnosis of clear cell ependymoma, and even the latest WHO classification does not shed light on that matter.12 Furthermore, they have to be distinguished from oligodendroglioma and central neurocytoma. Four recently published literature reviews and small series reported the clinical and immunohistochemical data of only about 50 cases,4, 5, 6, 7 while most specimens are likely to have been listed as ‘classic’ or ‘anaplastic’ ependymoma. Recent publications on the value of immunohistochemistry in the differential diagnosis of clear cell primary brain tumors6 and ependymomas13 have shown that clear cell ependymomas frequently show an expression of GFAP in the fibrillary matrix and a cytoplasmic punctate, dot-like positivity for EMA,6 the latter of which is highly specific and sensitive for ependymomas where it is encountered in 89% of cases.13 Thus, we were able to unequivocally identify the tumors in our series as being clear cell ependymomas.
Compared with other ependymomas, little is known regarding the molecular pathogenesis of the clear cell variant. So far, only one study has dealt with the genetic properties of a group of 10 clear cell ependymomas.5 By applying FISH, the authors found no detectable losses involving probes for NF2, 1p, or 19q, but demonstrated loss of the DAL-1 locus, a member of the protein 4.1 superfamily on 18p11.3, in 50% of their cases.5 In our cohort, however, no loss of DNA material from chromosome 18 was encountered; equally, no aberrations were found on 1p or 19q (Table 1). Instead, our CGH data revealed loss of chromosome 9 to be by far the most common DNA copy number change among clear cell ependymomas, accounting for 77% of all cases and involving 40% of WHO grade II and 100% of WHO grade III samples. In contrast, a recent extensive survey of CGH studies on pediatric brain tumors found loss of chromosome 9 to be absent in classic ependymomas (which—if at all—showed gains of chromosome 9 in 6–8% of cases), and to occur in only 13–16% of anaplastic ependymomas.10 This is corroborated by a meta-analysis of 509 gliomas investigated by CGH, in which +9 was listed as a characteristic molecular marker for ependymomas, in particular low-grade ependymomas,14 with additional changes consisting of +1q, +18, and −22q.15 A recent loss of heterozygosity (LOH) study saw a deletion of 9q in nine of 33 ependymomas, of which all bar two, however, were located infratentorially.16 Potential tumor suppressor genes on 9p are CDKN2B/p15, CDKN2A/p16, p14 ARF, and ABL1, whereas 9q contains the TSC1 and PTCH tumor suppressor genes. The former is implicated in the pathogenesis of subependymal giant cell astrocytomas in tuberous sclerosis complex (TSC), while the latter is mutated in sporadic and familial medulloblastomas associated with Gorlin's syndrome.17
The second most common imbalance in our cohort was +1q, which was exclusively encountered in the anaplastic clear cell variant where it was found in 63% of cases. This is above the rate of 30% usually found in anaplastic ependymomas by CGH where +1q is the most common chromosomal imbalance.10 Potential protooncogenes mapping to 1q are SKI, PBX1, TRK/TRKC, ABL2 and ELK4,9 and increased gene expression patterns have been found for PRELP, EPHX1, FY and HSPA6.18 Interestingly, classic ependymomas with +1q tended to occur in the posterior fossa and were associated with a poorer outcome regardless of tumor grade,19 whereas in our cohort four of five cases showing +1q were found in a supratentorial location and three of five patients—all of them with anaplastic ependymomas and high-level gains of 1q—were still alive after observation periods ranging from 23 to 38 months (Table 1). In the former study, +1q was strongly linked with tumor recurrence,19 a feature also found in four of five of our patients.
The third most frequent change consisted of loss of chromosome 3 in 31% of all clear cell ependymomas, an aberration that generally does not feature among ependymoma CGH.10 However, a recent LOH study found deletion of 3p in 13% of ependymomas.16 This region is intriguing, as the VHL (von Hippel-Lindau disease) tumor suppressor gene maps to 3p25–26 which is associated with the occurrence of hemangioblastomas, another tumor characterized by a clear-cell pattern.
The next most frequent aberration involved loss of 22q, which was found in 20% of WHO grade II and 25% of WHO grade III clear cell ependymomas (Table 1). While the former figure is in the order expected of classic ependymomas in childhood (mean 31%),10 the latter is much higher than among anaplastic ependymomas where −22q is rare and mainly found in infratentorial and spinal cases.10, 16 Potential tumor suppressor genes mapping to 22q include BAM22, hSNF5/INI1, and the neurofibromatosis type 2 (NF2) gene.10 In adults, allelic loss of chromosome arm 22q occurs in up to 60% of cases, with some showing an altered NF2 gene; involvement of hSNF5/INI1—usually associated with atypical teratoid/rhabdoid tumors—could be excluded.20 Loss of chromosome 22q is also frequently encountered among choroid plexus tumors, meningiomas, neurinomas, atypical teratoid/rhabdoid tumors, pediatric high-grade astrocytomas, and pineoblastomas.10 Of the less frequent gains of 4q and 13q found in our cohort of clear cell ependymomas, +4q has only been encountered in one CGH study of classic ependymomas of WHO grade II (nine of 20 cases),21 whereas +13q did not feature at all among other ependymoma variants.10
In contrast, a recent study of 33 ependymomas of grades I-III found LOH of 6q to be the most common deletion (33% of cases), with all but two of them located infratentorially,16 a finding reiterating earlier CGH data on ependymomas carrying a loss of 6q.22, 23 Interestingly, −6q was found in only one of our clear cell ependymomas which also happened to be the sole infratentorial tumor (case 12, Table 1).
Finally, in contrast to two previous CGH studies showing an inverse correlation between tumor grade and DNA copy number changes among ependymomas (WHO grade I: 9.1–16, WHO grade II: 5.3–12, and WHO grade III: 1.8–5 aberrations/tumor),14, 24 our clear cell ependymomas of WHO grade II showed fewer imbalances than WHO grade III ones with 1.4 vs 3.5 aberrations per tumor, respectively, incidentally also carrying altogether fewer alterations than usually recorded for ependymomas.10
In conclusion, we present a systematic CGH analysis of 13 almost exclusively supratentorial clear cell ependymomas. Although some of the implicated chromosomes have been shown to be involved in other ependymoma variants, the striking frequency of +1q and −9 (and less so of −3), which usually do not feature in ependymomas or if so, only in infratentorial cases, shows that aberrations differ between clear cell and other types of ependymomas.
References
Kawano N, Yada K, Aihara M, et al. Oligodendroglioma-like cells (clear cells) in ependymoma. Acta Neuropathol 1983;62:141–144.
Kawano N, Yada K, Yagishita S . Clear cell ependymoma. A histological variant with diagnostic implications. Virchows Arch A Pathol Anat Histopathol 1989;415:467–472.
Kaatsch P, Rickert CH, Kühl J, et al. Population-based epidemiological data of brain tumors in German children. Cancer 2001;92:3155–3164.
Amatya VJ, Takeshima Y, Kaneko M, et al. Case of clear cell ependymoma of medulla oblongata: clinicopathological and immunohistochemical study with literature review. Pathol Int 2003;53:297–302.
Fouladi M, Helton K, Dalton J, et al. Clear cell ependymoma: a clinicopathologic and radiographic analysis of 10 patients. Cancer 2003;98:2232–2244.
Koperek O, Gelpi E, Birner P, et al. Value and limits of immunohistochemistry in differential diagnosis of clear cell primary brain tumors. Acta Neuropathol 2004;108:24–30.
Suri VS, Tatke M, Singh D, et al. Histological spectrum of ependymomas and correlation of p53 and Ki-67 expression with ependymoma grade and subtype. Indian J Cancer 2004;41:66–71.
Min KW, Scheithauer BW . Clear cell ependymoma: a mimic of oligodendroglioma: clinicopathologic and ultrastructural considerations. Am J Surg Pathol 1997;21:820–826.
Rickert CH, Sträter R, Kaatsch P, et al. Pediatric high-grade astrocytomas show chromosomal imbalances distinct from adult cases. Am J Pathol 2001;158:1525–1532.
Rickert CH, Paulus W . Comparative genomic hybridization in central and peripheral nervous system tumors of childhood and adolescence. J Neuropathol Exp Neurol 2004;63:399–417.
Kleihues P, Burger PC, Scheithauer BW . The new WHO classification of brain tumours. Brain Pathol 1993;3:255–268.
Wiestler OD, Schiffer D, Coons SW, et al. Ependymoma. In: P Kleihues, WK Cavenee (eds). Pathology & Genetics. Tumours of the nervous system. WHO: Lyon, 2000, p 73.
Hasselblatt M, Paulus W . Sensitivity and specificity of epithelial membrane antigen staining patterns in ependymomas. Acta Neuropathol 2003;106:385–388.
Scheil S, Brüderlein S, Eickert M, et al. Low frequency of chromosomal imbalances in anaplastic ependymomas detected by comparative genomic hybridization. Brain Pathol 2001;11:133–143.
Koschny R, Koschny T, Froster UG, et al. Comparative genomic hybridization in glioma: a meta-analysis of 509 cases. Cancer Genet Cytogenet 2002;135:147–159.
Huang B, Starostik P, Schraut H, et al. Human ependymomas reveal frequent deletions on chromosomes 6 and 9. Acta Neuropathol 2003;106:357–362.
Pietsch T, Waha A, Koch A, et al. Medulloblastomas of the desmoplastic variant carry mutations of the human homologue of Drosophila patched. Cancer Res 1997;57:2085–2088.
Korshunov A, Neben K, Wrobel G, et al. Gene expression patterns in ependymomas correlate with tumor location, grade, and patient age. Am J Pathol 2003;163:1721–1727.
Dyer S, Prebble E, Davison V, et al. Genomic imbalances in pediatric intracranial ependymomas define clinically relevant groups. Am J Pathol 2002;161:2133–2141.
Kraus JA, de Millas W, Sörensen N, et al. Indications for a tumor suppressor gene at 22q11 involved in the pathogenesis of ependymal tumors and distinct from hSNF5/INI1. Acta Neuropathol 2001;102:69–74.
Jeuken JW, Sprenger SH, Gilhuis J, et al. Correlation between localization, age, and chromosomal imbalances in ependymal tumours as detected by CGH. J Pathol 2002;197:238–244.
Hirose Y, Aldape K, Bollen A, et al. Chromosomal abnormalities subdivide ependymal tumors into clinically relevant groups. Am J Pathol 2001;158:1137–1143.
Carter M, Nicholson J, Ross F, et al. Genetic abnormalities detected in ependymomas by comparative genomic hybridisation. Br J Cancer 2002;86:929–939.
Gilhuis HJ, van der Laak J, Wesseling P, et al. Inverse correlation between genetic aberrations and malignancy grade in ependymal tumors: a paradox? J Neurooncol 2004;66:111–116.
Acknowledgements
The invaluable help and skillful assistance of Beate Schröder is gratefully appreciated.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rickert, C., Korshunov, A. & Paulus, W. Chromosomal imbalances in clear cell ependymomas. Mod Pathol 19, 958–962 (2006). https://doi.org/10.1038/modpathol.3800614
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800614
Keywords
This article is cited by
-
Molecular characterization of histopathological ependymoma variants
Acta Neuropathologica (2020)
-
Clear cell ependymoma with trisomy 19 developing bone metastases
Child's Nervous System (2012)
-
A case of anaplastic clear-cell ependymoma presenting with high erythropoietin concentration and 1p/19q deletions
Brain Tumor Pathology (2011)
-
Trisomy 19 ependymoma, a newly recognized genetico-histological association, including clear cell ependymoma
Molecular Cancer (2007)
-
Molecular genetics of pediatric central nervous system tumors
Current Oncology Reports (2006)