Abstract
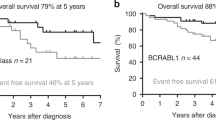
Minimal residual disease (MRD) monitoring is an essential tool for risk group stratification in current treatment protocols for childhood acute lymphoblastic leukaemia (ALL). Although quantitative detection of clonal immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements is currently considered to be the standard method, leukaemia fusion genes provide other possible targets for MRD follow-up, as already demonstrated in TEL/AML1-positive ALLs. We analysed and compared MRD levels quantified by BCR/ABL transcript detection and by the standard Ig/TCR-based method in 218 bone marrow specimens from 17 children with BCR/ABL-positive ALL. We found only a limited overall correlation of MRD levels as assessed by the two methods (correlation coefficient R2=0.64). The correlation varied among patients from excellent (R2=0.99) to very poor (R2=0.17). Despite identical sensitivity of the approaches, 20% of the samples were negative by the Ig/TCR approach whereas positive by the BCR/ABL method. We show that multilineage involvement is at least partly responsible for the discrepancy. Moreover, our data demonstrate that BCR/ABL monitoring enables better and earlier prediction of relapse compared to the standard Ig/TCR methodology. We conclude that BCR/ABL-based MRD monitoring of childhood ALL is a clinically relevant tool and should be performed in parallel with the standard Ig/TCR follow-up.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schlieben S, Borkhardt A, Reinisch I, Ritterbach J, Janssen JW, Ratei R et al. Incidence and clinical outcome of children with BCR/ABL-positive acute lymphoblastic leukemia (ALL). A prospective RT-PCR study based on 673 patients enrolled in the German pediatric multicenter therapy trials ALL-BFM-90 and CoALL-05-92. Leukemia 1996; 10: 957–963.
Deininger MW, Goldman JM, Melo JV . The molecular biology of chronic myeloid leukemia. Blood 2000; 96: 3343–3356.
Suryanarayan K, Hunger SP, Kohler S, Carroll AJ, Crist W, Link MP et al. Consistent involvement of the bcr gene by 9;22 breakpoints in pediatric acute leukemias. Blood 1991; 77: 324–330.
Arico M, Valsecchi MG, Camitta B, Schrappe M, Chessells J, Baruchel A et al. Outcome of treatment in children with Philadelphia chromosome-positive acute lymphoblastic leukemia. N Engl J Med 2000; 342: 998–1006.
Fletcher JA, Lynch EA, Kimball VM, Donnelly M, Tantravahi R, Sallan SE . Translocation (9;22) is associated with extremely poor prognosis in intensively treated children with acute lymphoblastic leukemia. Blood 1991; 77: 435–439.
Uckun FM, Nachman JB, Sather HN, Sensel MG, Kraft P, Steinherz PG et al. Clinical significance of Philadelphia chromosome positive pediatric acute lymphoblastic leukemia in the context of contemporary intensive therapies: a report from the Children's Cancer Group. Cancer 1998; 83: 2030–2039.
Cazzaniga G, Lanciotti M, Rossi V, Di Martino D, Arico M, Valsecchi MG et al. Prospective molecular monitoring of BCR/ABL transcript in children with Ph+ acute lymphoblastic leukaemia unravels differences in treatment response. Br J Haematol 2002; 119: 445–453.
Ribeiro RC, Broniscer A, Rivera GK, Hancock ML, Raimondi SC, Sandlund JT et al. Philadelphia chromosome-positive acute lymphoblastic leukemia in children: durable responses to chemotherapy associated with low initial white blood cell counts. Leukemia 1997; 11: 1493–1496.
Schrappe M, Arico M, Harbott J, Biondi A, Zimmermann M, Conter V et al. Philadelphia chromosome-positive (Ph+) childhood acute lymphoblastic leukemia: good initial steroid response allows early prediction of a favorable treatment outcome. Blood 1998; 92 (8): 2730–2741.
Biondi A, Valsecchi MG, Seriu T, D'Aniello E, Willemse MJ, Fasching K et al. Molecular detection of minimal residual disease is a strong predictive factor of relapse in childhood B-lineage acute lymphoblastic leukemia with medium risk features. A case control study of the International BFM study group. Leukemia 2000; 14: 1939–1943.
Eckert C, Biondi A, Seeger K, Cazzaniga G, Hartmann R, Beyermann B et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. Lancet 2001; 358: 1239–1241.
Panzer-Grumayer ER, Schneider M, Panzer S, Fasching K, Gadner H . Rapid molecular response during early induction chemotherapy predicts a good outcome in childhood acute lymphoblastic leukemia. Blood 2000; 95: 790–794.
Sramkova L, Muzikova K, Fronkova E, Krejci O, Sedlacek P, Formankova R et al. Detectable minimal residual disease before allogeneic hematopoietic stem cell transplantation predicts extremely poor prognosis in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 2007; 48: 93–100.
van der Velden VH, Joosten SA, Willemse MJ, van Wering ER, Lankester AW, van Dongen JJ et al. Real-time quantitative PCR for detection of minimal residual disease before allogeneic stem cell transplantation predicts outcome in children with acute lymphoblastic leukemia. Leukemia 2001; 15: 1485–1487.
van Dongen JJ, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998; 352: 1731–1738.
Krejci O, Prouzova Z, Horvath O, Trka J, Hrusak O . Cutting edge: TCR delta gene is frequently rearranged in adult B lymphocytes. J Immunol 2003; 171: 524–527.
van der Velden VH, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007; 21: 604–611.
van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Pongers-Willemse MJ, Seriu T, Stolz F, d'Aniello E, Gameiro P, Pisa P et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia. Leukemia 1999; 13: 110–118.
Baruchel A, Cayuela JM, MacIntyre E, Berger R, Sigaux F . Assessment of clonal evolution at Ig/TCR loci in acute lymphoblastic leukaemia by single-strand conformation polymorphism studies and highly resolutive PCR derived methods: implication for a general strategy of minimal residual disease detection. Br J Haematol 1995; 90: 85–93.
Beishuizen A, Verhoeven MA, van Wering ER, Hahlen K, Hooijkaas H, van Dongen JJ . Analysis of Ig and T-cell receptor genes in 40 childhood acute lymphoblastic leukemias at diagnosis and subsequent relapse: implications for the detection of minimal residual disease by polymerase chain reaction analysis. Blood 1994; 83: 2238–2247.
Steward CG, Goulden NJ, Katz F, Baines D, Martin PG, Langlands K et al. A polymerase chain reaction study of the stability of Ig heavy-chain and T-cell receptor delta gene rearrangements between presentation and relapse of childhood B-lineage acute lymphoblastic leukemia. Blood 1994; 83: 1355–1362.
Szczepanski T, Willemse MJ, Brinkhof B, van Wering ER, van der Burg M, van Dongen JJ . Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B-ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood 2002; 99: 2315–2323.
Szczepanski T . Why and how to quantify minimal residual disease in acute lymphoblastic leukemia? Leukemia 2007; 21: 622–626.
Fronkova E, Madzo J, Zuna J, Reznickova L, Muzikova K, Hrusak O et al. TEL/AML 1 real-time quantitative reverse transcriptase PCR can complement minimal residual disease assessment in childhood ALL. Leukemia 2005; 19: 1296–1297.
Madzo J, Zuna J, Muzikova K, Kalinova M, Krejci O, Hrusak O et al. Slower molecular response to treatment predicts poor outcome in patients with TEL/AML1 positive acute lymphoblastic leukemia: prospective real-time quantitative reverse transcriptase-polymerase chain reaction study. Cancer 2003; 97: 105–113.
Miller SA, Dykes DD, Polesky HF . A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Chomczynski P, Sacchi N . Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156–159.
Bruggemann M, van der Velden VH, Raff T, Droese J, Ritgen M, Pott C et al. Rearranged T-cell receptor beta genes represent powerful targets for quantification of minimal residual disease in childhood and adult T-cell acute lymphoblastic leukemia. Leukemia 2004; 18: 709–719.
Langerak AW, Wolvers-Tettero IL, van Gastel-Mol EJ, Oud ME, van Dongen JJ . Basic helix-loop-helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood 2001; 98: 2456–2465.
van der Velden VH, Wijkhuijs JM, Jacobs DC, van Wering ER, van Dongen JJ . T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia 2002; 16: 1372–1380.
van der Velden VH, Willemse MJ, van der Schoot CE, Hahlen K, van Wering ER, van Dongen JJ . Immunoglobulin kappa deleting element rearrangements in precursor-B acute lymphoblastic leukemia are stable targets for detection of minimal residual disease by real-time quantitative PCR. Leukemia 2002; 16: 928–936.
Verhagen OJ, Willemse MJ, Breunis WB, Wijkhuijs AJ, Jacobs DC, Joosten SA et al. Application of germline IGH probes in real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia. Leukemia 2000; 14: 1426–1435.
Pongers Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
Yokota H, Tsuno NH, Tanaka Y, Fukui T, Kitamura K, Hirai H et al. Quantification of minimal residual disease in patients with e1a2 BCR-ABL-positive acute lymphoblastic leukemia using a real-time RT-PCR assay. Leukemia 2002; 16: 1167–1175.
Elmaagacli AH, Beelen DW, Opalka B, Seeber S, Schaefer UW . The amount of BCR-ABL fusion transcripts detected by the real-time quantitative polymerase chain reaction method in patients with Philadelphia chromosome positive chronic myeloid leukemia correlates with the disease stage. Ann Hematol 2000; 79: 424–431.
Gaiger A, Henn T, Horth E, Geissler K, Mitterbauer G, Maier-Dobersberger T et al. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood 1995; 86: 2371–2378.
van Rhee F, Hochhaus A, Lin F, Melo JV, Goldman JM, Cross NC . p190 BCR-ABL mRNA is expressed at low levels in p210-positive chronic myeloid and acute lymphoblastic leukemias. Blood 1996; 87: 5213–5217.
Fuster JL, Bermudez M, Galera A, Llinares ME, Calle D, Ortuno FJ . Imatinib mesylate in combination with chemotherapy in four children with de novo and advanced stage Philadelphia chromosome-positive acute lymphoblastic leukemia. Haematologica 2007; 92: 1723–1724.
Dow LW, Tachibana N, Raimondi SC, Lauer SJ, Witte ON, Clark SS . Comparative biochemical and cytogenetic studies of childhood acute lymphoblastic leukemia with the Philadelphia chromosome and other 22q 11 variants. Blood 1989; 73: 1291–1297.
Estrov Z, Talpaz M, Kantarjian HM, Zipf TF, McClain KL, Kurzrock R . Heterogeneity in lineage derivation of Philadelphia-positive acute lymphoblastic leukemia expressing p190BCR-ABL or p210BCR-ABL: determination by analysis of individual colonies with the polymerase chain reaction. Cancer Res 1993; 53: 3289–3293.
Primo D, Sanchez ML, Espinosa AB, Tabernero MD, Rasillo A, Sayagues JM et al. Lineage involvement in chronic myeloid leukaemia: comparison between MBCR/ABL and mBCR/ABL cases. Br J Haematol 2006; 132: 736–739.
Secker-Walker LM, Cooke HM, Browett PJ, Shippey CA, Norton JD, Coustan-Smith E et al. Variable Philadelphia breakpoints and potential lineage restriction of bcr rearrangement in acute lymphoblastic leukemia. Blood 1988; 72: 784–791.
Schenk TM, Keyhani A, Bottcher S, Kliche KO, Goodacre A, Guo JQ et al. Multilineage involvement of Philadelphia chromosome positive acute lymphoblastic leukemia. Leukemia 1998; 12: 666–674.
Tachibana N, Raimondi SC, Lauer SJ, Sartain P, Dow LW . Evidence for a multipotential stem cell disease in some childhood Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 1987; 70: 1458–1461.
Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia 2007; 21: 926–935.
Wetzler M, Talpaz M, Van Etten RA, Hirsh-Ginsberg C, Beran M, Kurzrock R . Subcellular localization of Bcr, Abl, and Bcr-Abl proteins in normal and leukemic cells and correlation of expression with myeloid differentiation. J Clin Invest 1993; 92: 1925–1939.
Acknowledgements
This work was supported by grants MSM0021620813, MZO00064203 and NS10004. We acknowledge all participating centres within the Czech Paediatric Haematology Working Group (CPH) for clinical management and sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zaliova, M., Fronkova, E., Krejcikova, K. et al. Quantification of fusion transcript reveals a subgroup with distinct biological properties and predicts relapse in BCR/ABL-positive ALL: implications for residual disease monitoring. Leukemia 23, 944–951 (2009). https://doi.org/10.1038/leu.2008.386
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2008.386
Keywords
This article is cited by
-
Minimal residual disease in BCR::ABL1-positive acute lymphoblastic leukemia: different significance in typical ALL and in CML-like disease
Leukemia (2022)
-
Standardisation and consensus guidelines for minimal residual disease assessment in Philadelphia-positive acute lymphoblastic leukemia (Ph + ALL) by real-time quantitative reverse transcriptase PCR of e1a2 BCR-ABL1
Leukemia (2019)
-
Role of minimal residual disease in the management of acute myeloid leukemia—a case-based discussion
Annals of Hematology (2018)
-
Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL
Bone Marrow Transplantation (2017)
-
Characterization of 46 patient-specific BCR-ABL1 fusions and detection of SNPs upstream and downstream the breakpoints in chronic myeloid leukemia using next generation sequencing
Molecular Cancer (2015)