Abstract
The autosomal dominant macrothrombocytopenia with leukocyte inclusions, May-Hegglin anomaly, Sebastian syndrome, and Fechtner syndrome, are rare human disorders characterized by a triad of giant platelets, thrombocytopenia, and characteristic Döhle body-like cytoplasmic inclusions in granulocytes. Epstein syndrome is another autosomal dominant macrothrombocytopenia associated with Alport syndrome but without leukocyte inclusions. These disorders are caused by mutations in the same gene, the MYH9, which encodes the nonmuscle myosin heavy chain-A (NMMHCA). The term, MYH9 disorders, has been proposed, but the clinicopathologic basis of MYH9 mutations has been poorly investigated. In this study, a total of 24 cases with MYH9 disorders and suspected cases were subjected to immunofluorescence analysis by a polyclonal antibody against human platelet NMMHCA. Abnormal subcellular localization of NMMHCA was observed in every neutrophil from individuals with MYH9 mutations. Comparison with May-Grünwald-Giemsa staining revealed that the NMMHCA always coexisted with the neutrophil inclusion bodies, suggesting that NMMHCA is associated with such bodies. In three cases, neutrophil inclusions were not detected on conventional May-Grünwald-Giemsa-stained blood smears but immunofluorescence analysis revealed the abnormal NMMHCA localization. In contrast, cases with Epstein syndrome and the isolated macrothrombocytopenia with normal NMMHCA localization had no MYH9 mutations. An antibody that recognizes the C-terminal 12 mer peptides showed similar immunoreactivity from the patients heterozygous for truncated mutations that abolished the C-terminal epitope, suggesting that normal NMMHCA dimerizes with abnormal NMMHCA to form inclusion bodies. We further propose that the localization pattern can be classified into three groups according to the number, size, and shape of the fluorescence-labeled NMMHCA granule. Immunofluorescence analysis of neutrophil NMMHCA is useful as a screening test for the clear hematopathologic classification of MYH9 disorders.
Similar content being viewed by others
Introduction
The autosomal dominant macrothrombocytopenia with leukocyte inclusions, May-Hegglin anomaly (MHA; MIM 155100) (Hegglin, 1945; May, 1909), Sebastian syndrome (SBS; MIM 605249) (Greinacher et al, 1990), and Fechtner syndrome (FTNS; MIM 153640) (Peterson et al, 1985), are rare human disorders characterized by a triad of giant platelets, thrombocytopenia, and characteristic Döhle body-like cytoplasmic inclusions in granulocytes. FTNS is associated with a wide range of Alport syndrome symptoms, including nephritis, deafness, and cataracts, whereas Epstein syndrome (EPS; MIM 153650) is another autosomal dominant macrothrombocytopenia associated with Alport syndrome but without leukocyte inclusions (Epstein et al, 1972). By a positional cloning approach, we and others have recently established that the MYH9, the gene encoding the nonmuscle myosin heavy chain-A (NMMHCA), is mutated in these disorders (Kelley et al, 2000; Kunishima et al, 2001a; The May-Hegglin/Fechtner Syndrome Consortium, 2000). Several MYH9 mutations have been documented among patients, including those with a nonsyndromic deafness, DFNA17 (MIM 603622) (Lalwani et al, 2000), and these disorders appear to represent a class of allelic disorders (Arrondel et al, 2002; Heath et al, 2001; Kunishima et al, 2001b). Accordingly, the term MYH9 (or NMMHCA) disorders has been proposed. In addition to these characteristic macrothrombocytopenias, there is an isolated or nonsyndromic form, which is not associated with leukocyte inclusions nor with other abnormalities. It is not known whether defects in MYH9 contribute to the etiology of isolated macrothrombocytopenia.
The diagnosis of MHA, SBS, and FTNS has been conventionally made on the basis of hematomorphologic examinations detecting a triad of giant platelets, thrombocytopenia, and inclusion bodies in the cytoplasm of granulocytes (Hegglin, 1945; May, 1909). Inclusion bodies are usually identified on May-Grünwald-Giemsa (MGG) or Wright’s stained blood smears. Although the hallmark of these disorders is the presence of leukocyte inclusions, their faint appearance occasionally hampers proper recognition. We have identified an abnormal localization of NMMHCA in MHA and SBS neutrophils by immunofluorescence analysis and suggested the usefulness of this method for the diagnosis of these disorders (Kunishima et al, 2001a). In this study, we further investigated the neutrophil NMMHCA localization in MYH9 disorders by comparing the immunoreactivity pattern and the mutational status of the MYH9 gene.
Results
Localization of Neutrophil NMMHCA in MYH9 Disorders
Immunofluorescence analysis of neutrophil NMMHCA was performed using the rabbit polyclonal antibody against human platelet NMMHCA. In agreement with previous studies, normal neutrophils displayed a diffuse homogeneous distribution of NMMHCA throughout the cytoplasm and sometimes in perinucleus (Fig. 1, B and D) (Kunishima et al, 2001a; Maupin et al, 1994). Analysis of neutrophil NMMHCA in patients with MYH9 disorders revealed a loss of homogeneous cytosolic distribution. Representative results are shown in Figures 2 to 4. The abnormal subcellular localization of NMMHCA was observed in every neutrophil as punctuate structures of various number and size. In each patient, staining patterns—including their number, size, and shape—were uniform and similar to those of the cytoplasmic inclusions observed on MGG-stained blood smears. Normal NMMHCA distribution in patients’ lymphocytes is consistent with the finding that inclusion bodies are not found in lymphocytes (data not shown).
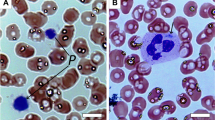
Nonmuscle myosin heavy chain-A (NMMHCA) localization in normal neutrophils. A and C, Light micrographs of May-Grünwald-Giemsa-stained neutrophils from two normal individuals. B and D, Immunofluorescence micrographs of neutrophils immunostained with the antiplatelet NMMHCA antibody. In normal neutrophils, NMMHCA is homogeneously distributed in the cytoplasm. Original magnification, ×1000.
Type I localization pattern of neutrophil NMMHCA in MYH9 disorders. Peripheral blood smears from case 14 (E1841K) (A, B), case 16 (5779delC) (C–E), case 21 (5818delG) (F–H), and case 17 (R1933X) (I–K) were stained with May-Grünwald-Giemsa (A, C, F, I), antiplatelet NMMHCA antibody (B, D, G, J), or anti-NMMHCA peptide antibody (E, H, K). Inclusion bodies in neutrophils are indicated by arrowheads. Original magnification, ×1000.
Type III localization pattern of neutrophil NMMHCA in MYH9 disorders. Peripheral blood smears from case 1 (N93K) (A, B), case 2 (S96L) (C, D), and case 11 (I1816V) (E, F) were stained with May-Grünwald-Giemsa (A, C, E) or anti-platelet NMMHCA antibody (B, D, F). Neutrophils from these cases contain no inclusion bodies. Original magnification, ×1000.
A different anti-NMMHCA antibody that recognizes the C-terminal 12 mer peptide sequences was also used in the study. A similar immunoreactivity of NMMHCA with the antihuman platelet NMMHCA antibody and the antipeptide antibody was consistently observed (Fig. 2, D and E, G and H, and J and K, respectively). Mutant NMMHCA polypeptides associated with two deletion mutations (5779delC and 5818delG) or a nonsense mutation (R1933X) in exon 40 do not have an epitope recognized by the antipeptide antibody and thus do not react with those truncated polypeptides within the cells. However, the similar immunoreactivity obtained by the two antibodies suggests that normal NMMHCA associates with abnormal NMMHCA to form inclusion bodies.
NMMHCA Accumulates at Inclusion Bodies
To obtain additional information about the relationship between MGG-stained inclusion bodies and NMMHCA localization, we sequentially treated peripheral blood smears with MGG staining and immunofluorescence analysis with the antiplatelet NMMHCA antibody. To do this, we first photographed MGG-stained neutrophils that contained inclusion bodies. Then we immunostained the cells in the same slide with the anti-NMMHCA antibody and analyzed the marked position by immunofluorescence microscopy. As shown in Figure 5, NMMHCA-positive fluorescent granules exactly coincided with the blue inclusion bodies stained with MGG, demonstrating that accumulated NMMHCA is responsible for the morphological appearance of inclusion bodies.
Identical localization of inclusion bodies and NMMHCA. A peripheral blood smear from case 15 with E1841K was sequentially stained with May-Grünwald-Giemsa (A, C) and antiplatelet NMMHCA antibody (B, D). A consistent appearance of inclusion bodies and NMMHCA-positive fluorescence was observed in two neutrophils (A/B, C/D). Inclusion bodies and NMMHCA-positive fluorescence granules are indicated by arrowheads. Original magnification, ×1000.
Localization Pattern of Neutrophil NMMHCA Classification into Three Groups
According to the morphology of immunoreactivity of neutrophil NMMHCA, three different localization patterns were identified in patients with MYH9 disorders (Figs. 2 to 4; Table 1). In type I, neutrophils contained one or two large, intensely stained, oval- to spindle-shaped cytoplasmic foci and low background (Fig. 2, B, D, E, G, H, J, and K). In this group, inclusion bodies on MGG-stained blood smears were mostly blue and spindle shaped with a diameter of approximately 0.5 to 2 μm (Fig. 2, A, C, F, and I). Type I includes four cases with E1841K mutation in exon 38 and six cases with mutations in the carboxyl-terminal domain in the last exon of 40: four R1933X and two deletion mutations that result in premature termination. Study of NMMHCA mutation further suggested that neutrophils from individuals with E1841K and deletion mutations had exclusively one or two large and intensely stained foci, whereas those from R1933X exhibited additional small dots (Fig. 2, J and K).
Type II NMMHCA localization included 8 cases in which neutrophils contained 3 to 20 circular to oval-shaped cytoplasmic punctuate spots with a diameter less than 1 μm (Fig. 3, B, D, F, and H). Individuals with type II localization were associated with mutations within exons 26 (R1165C, R1165L, and del L1205-Q1207) and exon 30 (D1424H, D1424N, and D1424Y).
Type II localization pattern of neutrophil NMMHCA in MYH9 disorders. Peripheral blood smears from case 3 (R1165C) (A, B), case 7 (delL1205-Q1207) (C, D), case 9 (D1424N) (E, F), and case 10 (D1424Y) (G, H) were stained with May-Grünwald-Giemsa (A, C, E, G) or antiplatelet NMMHCA antibody (B, D, F, H). Inclusion bodies in neutrophils are indicated by arrowheads. Original magnification, ×1000.
In the type III group, neutrophil inclusions were not detected on MGG stained blood smears (Fig. 4, A, C, and E), and indeed three patients have been diagnosed with EPS or isolated macrothrombocytopenia. However, the pattern of NMMHCA localization was abnormal, showing a speckled pattern or small dots scattered throughout the cytoplasm (Fig. 4, B, D, and F). In each case with type III localization, missense mutations leading to amino acid substitutions were detected, N93K and S96L in exon 1 and I1816V in exon 37. N93K and S96L, detected in an isolated macrothrombocytopenia in this study, have been reported in a MHA/SBS patient and a FTNS/EPS or a EPS patient, respectively (Arrondel et al, 2002; The May-Hegglin/Fechtner Syndrome Consortium, 2000). I1816V, the novel and first mutation detected in exon 37, was identified in a patient with EPS. This mutation cosegregated with the EPS phenotype in the family. These mutations were screened in 50 normal individuals, and none was found (data not shown).
Three cases had normal distribution of NMMHCA (Fig. 6, B and D), and the clinicopathological manifestation indicated that patient 22 had EPS, and patients 23 and 24 had isolated macrothrombocytopenia. In all these cases, no MYH9 mutations were found by a complete mutational screening of the entire 40 coding exons, suggesting that the abnormal NMMHCA localization is dependent on MYH9 mutation.
Normal localization of neutrophil NMMHCA from Epstein syndrome and isolated macrothrombocytopenia without MYH9 mutations. Peripheral blood smears from case 22 (A, B) and case 23 (C, D) were stained with May-Grünwald-Giemsa (A, C) or antiplatelet NMMHCA antibody (B, D). Original magnification, ×1000.
Discussion
A variety of clinical spectra of phenotypes caused by MYH9 mutations, frequently present in various combinations, might reflect the diverse functions of NMMHCA protein and the different influences of MYH9 mutations on the function of this motor protein in affected tissues. Alternatively, different properties of NMMHCA mutants or defects in tissue-specific functional domains may explain the diversity. Previous studies on genetic analysis of MYH9 have failed to display a clear relationship between clinical phenotypes and the sites of the MYH9 mutations (Arrondel et al, 2002; Heath et al, 2001; Kunishima et al, 2001b). For example, the clinical phenotype of individuals sharing the same mutation is variable. First, a mutation, D1424N, originally described in a large Japanese family with MHA, has been found in patients with SBS, EPS, and FTNS (Arrondel et al, 2002; Heath et al, 2001; Kunishima et al, 2001a). A nonsense mutation at R1933 has resulted in MHA, SBS, and FTNS (Arrondel et al, 2002; Heath et al, 2001; Kunishima et al, 2001a). Second, the expression of Alport manifestation in individuals sharing the same mutation is inconsistent even within a same family (Peterson et al, 1985; Rocca et al, 1993). The variability in clinical phenotypes and penetrance suggests that unknown genetic or epigenetic factors might influence the phenotypic consequences of MYH9 mutations. Although the molecular mechanisms of the influence by abnormal NMMHCA in affected tissues need to be clarified, difficulties in the availability of affected tissue samples such as kidney or inner ear hamper further histopathologic investigation. Peripheral blood neutrophils, in which characteristic cytoplasmic inclusion bodies are present in affected individuals, are easily available and thus suitable for hematopathological investigation. We have found abnormal localization of neutrophil NMMHCA in MHA and SBS patients, providing the first evidence that mutations of the MYH9 are related to the formation of leukocyte inclusions (Kunishima et al, 2001a). Our findings were recently confirmed by another group with immunohistochemical examination (Pecci et al, 2002). However, both studies did not show the correct subcellular localization of NMMHCA, and the possible correlation between mutational status of the MYH9 gene and localization pattern of NMMHCA has not yet been examined.
In the present study, abnormal subcellular localization of NMMHCA was observed in every neutrophil from patients with MYH9 disorders (Figs. 2 to 4). Also, the overall distribution of NMMHCA correlated well with that of inclusion bodies observed on MGG-stained blood smears. Furthermore, NMMHCA consistently accumulated at the site of inclusion bodies as revealed by sequential staining with MGG and NMMHCA immunostaining (Fig. 5). The simplest interpretation of these results may be that abnormal NMMHCA aggregates and accumulates in the cytoplasm. Previous electron microscopic studies have demonstrated that the inclusion bodies consist of ribosomes and microfilaments (Jenis et al, 1971; Lusher et al, 1968). It is thus conceivable that accumulated NMMHCA interacts with ribosomes or microfilaments and that such complex is visible as inclusion bodies on MGG-stained smears. Further experiments are required to determine the interaction between NMMHCA and ribosomes/microfilaments.
Myosins are the family of hexameric proteins composed of two heavy chains and two pairs of light chains, and they are involved in the motor function for diverse movements such as cytokinesis, cell shape, and specialized functions such as secretion and capping (Sellers, 2000). NMMHCA consists of both an N-terminal motor/head domain, harboring ATP- or actin-binding functions, and a C-terminal rod/tail domain that undergoes homodimerization to form a coiled-coil rod structure. In MYH9 disorders, the majority of the mutations have been identified in the rod domain, clustering in exons 25, 26, 30, 38, and 40. Mutations in the rod domain might result in disruption of dimerization or in loss of associations with other unknown proteins. In that case, the function of the head domain may not be affected completely. Indeed, 29 amino acid sequences in the C-terminal rod of the fast IId skeletal myosin are necessary for the assembly into thick myosin filaments (Sohn et al, 1997). Figure 2 indicates similar immunoreactivity in neutrophils from individuals with MYH9 truncation mutations, either by the polyclonal anti-NMMHCA antibody or the anti-NMMHCA C-terminal peptide antibody (Fig. 2, D and E, G and H, and J and K, respectively). Therefore, the wild-type protein participates in the inclusion body formation, suggesting that the interaction of wild-type and mutant proteins would produce dominant negative effects. This finding is quite consistent with the fact that all MYH9 disorders are inherited with autosomally dominant nature.
The localization pattern of neutrophil NMMHCA in MYH9 disorders can be classified into three groups according to their number, size, and shape, so we termed them type I, II, and III. The most characteristic feature was type I in which NMMHCA formed one or two large and intensely stained cytoplasmic foci by four of the MYH9 mutations (E1841K, R1933X, 5779delC, and 5818delG). Exclusively, one or two large foci were observed in E1841K, 5779delC, and 5818delG. So far, no patients with E1841K have been diagnosed with SBS (Heath et al, 2001; Kelley et al, 2000; Kunishima et al, 2001a, 2001b; The May-Hegglin/Fechtner Syndrome Consortium, 2000). R1933X, however, exhibited additional small dots, and two patients with this mutation were diagnosed with MHA and the other two with SBS in this study. From the previous reports, six patients with R1933X have been diagnosed with MHA and two with SBS (Heath et al, 2001; Kelley et al, 2000; Kunishima et al, 2001a, 2001b; The May-Hegglin/Fechtner Syndrome Consortium, 2000). The inclusions in MHA and SBS are different in size (MHA > SBS), shape (MHA oval to spindle, SBS round), and stainability (MHA > SBS) (Greinacher et al, 1990). Electron microscopic analysis can differentiate MHA from SBS by subtle ultrastructural features; in MHA, inclusions consist of ribosomes aligned along parallel microfilaments, whereas in SBS, the inclusions lack parallel filaments but are full of depolymerized ribosomes (Greinacher et al, 1990). In electron microscopic studies, inclusion bodies are often difficult to detect in ultrathin section, and if a small inclusion is present in a section, it is presumed to be an SBS-type inclusion. Therefore, the presence of additional small dots might affect the hematologic diagnosis of patients with R1933X mutation, even when a single granulocyte contains one or two large inclusions on MGG-stained blood smears. Type II neutrophils, consisting of several cytoplasmic spots with circular to oval shape, are associated with mutations within either exons 26 (R1165C, R1165L, and 1205_1207del) or exon 30 (D1424N, D1424H, and D1424Y). All the patients with exon 26 mutations were diagnosed with SBS, whereas those with exon 30 mutations were diagnosed with MHA. This may be because of the presence of larger NMMHCA-positive granules with exon 30 mutations rather than exon 26 mutations. However, further studies are required to interpret the size of immunostainable NMMHCA dots.
Hereditary macrothrombocytopenia is a genetically heterogeneous disorder, and the molecular mechanisms responsible for the production of giant platelets are not known (Mhawech and Saleem, 2000). If additional characteristic clinical or laboratory findings—such as abnormal platelet morphology, a bleeding tendency disproportionate to the platelet counts, or other associated abnormalities—are evident, an appropriate diagnosis can be made (Mhawech and Saleem, 2000). In this regard, detection of leukocyte inclusions, another characteristic feature and the conventional diagnostic criterion since the first description by May in 1909, is a necessary prerequisite for a proper diagnosis of MHA and SBS (May, 1909). Special attention is needed to identify the inclusion bodies because they may not be visible in all granulocytes, and inclusion bodies tend to become gradually less stained after blood sampling. Difficulty in discovery of inclusion bodies may sometimes lead to misdiagnosis of this disorder, such as a diagnosis of idiopathic thrombocytopenic purpura. Our current immunofluorescence technique represents a sensitive and accurate alternative to MGG staining in the detection and classification of the leukocyte inclusions. In particular, our technique successfully detected an abnormal speckled staining in neutrophils from patients with EPS and isolated macrothrombocytopenia associated with MYH9 mutations in which MGG-stained inclusion bodies have never been identified (Fig. 4). From the first description of EPS as a variant form of Alport syndrome, EPS has been characterized as having no leukocyte inclusion bodies (Eckstein et al, 1975; Epstein et al, 1972). However, our analysis clearly demonstrated that it is merely unrecognizable on standard MGG-stained blood smears. Moreover, results also suggest a genetic heterogeneity for EPS. Indeed, all three cases with EPS and the isolated macrothrombocytopenia with normal distribution of NMMHCA had no MYH9 mutations.
In conclusion, immunofluorescence analysis of neutrophil NMMHCA is useful for screening macrothrombocytopenias and in the hematopathologic classification of MYH9 disorders. It does not require experience in histopathology and can be performed on stored blood smears. Finally, the present investigation further broadens the spectrum of clinical phenotypes associated with MYH9 defects from the previously described six clinical phenotypes, MHA, SBS, FTNS, EPS, Alport-like syndromes, and DFNA17, to an isolated macrothrombocytopenia.
Materials and Methods
Patients and Samples
To investigate possible association between the pattern of subcellular localization of neutrophil NMMHCA protein and the location of the MYH9 mutation, we examined 24 samples of peripheral blood smears from patients with MYH9 disorders in which differential diagnoses of MHA and SBS have been confirmed by electron microscopic analysis and from suspected cases (Table 2). Control subjects consisted of a patient with Bernard-Soulier syndrome caused by a homozygous nonsense mutation of the glycoprotein Ibα gene (Kunishima et al, 1994) and several unaffected individuals from the families or sporadic cases. Peripheral blood was obtained after each individual gave informed consent for the study and the study had been approved by the institutional ethical committee.
Immunofluorescence Analysis of Leukocyte NMMHCA
The blood smears from all patients were used for the microscopic immunofluorescence analysis as described previously (Kunishima et al, 2001a). Briefly, peripheral blood samples were smeared on glass slides and air-dried. After being permeabilized with acetone for 1 minute at −20° C, the cells were hydrated and blocked with 10% normal swine serum for 10 minutes. Slides were incubated with antihuman platelet NMMHCA polyclonal antibody (BT561; Biomedical Technologies, Stoughton, Massachusetts) at 1:200 dilution and then reacted with rhodamine-labeled swine antirabbit IgG (DAKO, Glostrup, Denmark). The stained cells were examined under a fluorescence microscope (Eclipse E800; Nikon, Tokyo, Japan). In some experiments, rabbit polyclonal antibody raised against the C-terminal peptide sequences of human NMMHCA (GKADGAEAKPAE, 1950-1961aa; Berkeley Antibody Company, Richmond, California) was used. As negative controls, slides were incubated with normal rabbit serum instead of the primary antibody. In each staining experiment, normal control samples were simultaneously examined.
DNA Analysis of MYH9
Mutational analysis of MYH9 was performed as we described previously (Kunishima et al, 2001b). First, to screen for known MYH9 mutations, exons 1, 16, 25, 26, 30, 38, and 40 from genomic DNA from the patients were amplified by PCR. All amplified PCR fragments were subjected to restriction fragment length polymorphism analysis. If none of the reported mutations were detected, direct sequencing of the entire 40 coding exons and exon-intron boundaries of MYH9 were performed. All mutations were verified by reamplifying from genomic DNA and then repeating sequencing. The MYH9 cDNA sequence was numbered with the first ATG as +1, and amino acid residues are numbered from the ATG initiation codon (residue 1) (Saez et al, 1990; Toothaker et al, 1991).
References
Arrondel C, Vodovar N, Knebelmann B, Grunfeld JP, Gubler MC, Antignac C, and Heidet L (2002). Expression of the nonmuscle myosin heavy chain IIA in the human kidney and screening for MYH9 mutations in Epstein and Fechtner syndromes. J Am Soc Nephrol 13: 65–74.
Eckstein JD, Filip DJ, and Watts JC (1975). Hereditary thrombocytopenia, deafness and renal disease. Ann Intern Med 82: 639–645.
Epstein CJ, Sahud MA, Piel CF, Goodman JR, Bernfield MR, Kushner JH, and Ablin AR (1972). Hereditary macrothrombocytopathia, nephritis and deafness. Am J Med 52: 299–310.
Greinacher A, Nieuwenhuis HK, and White JG (1990). Sebastian platelet syndrome: A new variant of hereditary macrothrombocytopenia with leukocyte inclusions. Blut 61: 282–288.
Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, Denison JC, Gregory MC, White JG, Barker DF, Greinacher A, Epstein CJ, Glucksman MJ, and Martignetti JA (2001). Nonmuscle myosin heavy chain IIa mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet 69: 1033–1045.
Hegglin R (1945). Gleichzeitige konstitutionelle Veranderungen an Neutrophilen und Thrombocyten. Helv Med Acta 12: 439–440.
Jenis EH, Takeuchi A, Dillon DE, Ruymann FB, and Rivkin S (1971). The May-Hegglin anomaly: Ultrastructure of the granulocytic inclusion. Am J Clin Pathol 55: 187–196.
Kelley MJ, Jawien W, Ortel TL, and Korczak JF (2000). Mutation of MYH9, encoding non-muscle myosin heavy chain A, in May-Hegglin anomaly. Nat Genet 26: 106–108.
Kunishima S, Kojima T, Matsushita T, Tanaka T, Tsurusawa M, Furukawa Y, Nakamura Y, Okamura T, Amemiya N, Nakayama T, Kamiya T, and Saito H (2001a). Mutations in the NMMHC-A gene cause autosomal dominant macrothrombocytopenia with leukocyte inclusions (May-Hegglin anomaly/Sebastian syndrome). Blood 97: 1147–1149.
Kunishima S, Matsushita T, Kojima T, Amemiya N, Choi YM, Hosaka N, Inoue M, Jung Y, Mamiya S, Matsumoto K, Miyajima Y, Ruan C, Saito K, Song KS, Yoon HJ, Kamiya T, and Saito H (2001b). Identification of six novel MYH9 mutations and genotype-phenotype relationships in autosomal dominant macrothrombocytopenia with leukocyte inclusions. J Hum Genet 46: 722–729.
Kunishima S, Miura H, Fukutani H, Yoshida H, Osumi K, Kobayashi S, Ohno R, and Naoe T (1994). Bernard-Soulier syndrome Kagoshima: Ser 444–>stop mutation of glycoprotein (GP) Ibα resulting in circulating truncated GPIbα and surface expression of GPIbβ and GPIX. Blood 84: 3356–3362.
Lalwani AK, Golstein JA, Kelley MJ, Luxford W, Castelein CM, and Mhatre AN (2000). Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet 67: 1121–1128.
Lusher JM, Schneider J, Mizukami J, and Evans RK (1968). The May-Hegglin anomaly: Platelet function, ultrastructure and chromosome studies. Blood 32: 950–961.
Maupin P, Phillips CL, Adelstein RS, and Pollard TD (1994). Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci 107: 3077–3090.
May R (1909). Leukozyteneinschlusse. Dtsch Arch Klin Med 96: 1–6.
Mhawech P and Saleem A (2000). Inherited giant platelet disorders. Classification and literature review. Am J Clin Pathol 113: 176–190.
Pecci A, Noris P, Invernizzi R, Savoia A, Seri M, Ghiggeri GM, Sartore S, Gangarossa S, Bizzaro N, and Balduini CL (2002). Immunocytochemistry for the heavy chain of the non-muscle myosin IIA as a diagnostic tool for MYH9-related disorders. Br J Haematol 117: 164–167.
Peterson LC, Rao KV, Crosson JT, and White JG (1985). Fechtner syndrome: A variant of Alport’s syndrome with leukocyte inclusions and macrothrombocytopenia. Blood 65: 397–406.
Rocca B, Laghi F, Zini G, Maggiano N, and Landolfi R (1993). Fechtner syndrome: Report of a third family and literature review. Br J Haematol 85: 423–426.
Saez CG, Myers JC, Shows TB, and Leinwand LA (1990). Human nonmuscle myosin heavy chain mRNA: Generation of diversity through alternative polyadenylylation. Proc Natl Acad Sci USA 87: 1164–1168.
Sellers JR (2000). Myosins: A diverse superfamily. Biochim Biophys Acta 1496: 3–22.
Sohn RL, Vikstrom KL, Strauss M, Cohen C, Szent-Gyorgyi AG, and Leinwand LA (1997). A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol 266: 317–330.
The May-Hegglin/Fechtner Syndrome Consortium (2000). Mutations in MYH9 result in the May-Hegglin anomaly, and Fechtner and Sebastian syndromes. Nat Genet 26: 103–105.
Toothaker LE, Gonzalez DA, Tung N, Lemons RS, Le Beau MM, Arnaout MA, Clayton LK, and Tenen DG (1991). Cellular myosin heavy chain in human leukocytes: Isolation of 5′ cDNA clones, characterization of the protein, chromosomal localization, and upregulation during myeloid differentiation. Blood 78: 1826–1833.
Acknowledgements
This study was supported by Grants-in Aid for Scientific Research (10557090, 11470209, and 12670981) from the Ministry of Education, Culture, Sports, Science and Technology, and for Research on Specific Diseases from the Ministry of Health, Labor and Welfare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kunishima, S., Matsushita, T., Kojima, T. et al. Immunofluorescence Analysis of Neutrophil Nonmuscle Myosin Heavy Chain-A in MYH9 Disorders: Association of Subcellular Localization with MYH9 Mutations. Lab Invest 83, 115–122 (2003). https://doi.org/10.1097/01.LAB.0000050960.48774.17
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.LAB.0000050960.48774.17
This article is cited by
-
Further classification of neutrophil non-muscle myosin heavy chain-IIA localization for efficient genetic diagnosis of MYH9 disorders
Annals of Hematology (2018)
-
Ripor2 is involved in auditory hair cell stereociliary bundle structure and orientation
Journal of Molecular Medicine (2018)
-
Anesthetic management without perioperative platelet transfusion for cervical laminectomy and laminoplasty in a case of May–Hegglin anomaly
Journal of Anesthesia (2018)
-
Efficacy of neutrophil non-muscle myosin heavy chain-IIA immunofluorescence analysis in determining the pathogenicity of MYH9 variants
Annals of Hematology (2017)
-
A Japanese pedigree with a p.A95V mutation in the MYH9 gene demonstrates inherited macrothrombocytopenia without Alport manifestations
Annals of Hematology (2016)