Abstract
It is suggested that chromosome 18p11 is a susceptibility region for both bipolar disorder and schizophrenia. Aiming to identify susceptibility gene(s), we investigated a family whose members have either schizophrenia or schizophrenia-spectrum psychosis and carried a t(18;21)(p11.1;p11.1) translocation. Fluorescence in situ hybridization showed that the breakpoint on chromosome 21 was localized to a bacterial artificial chromosome (BAC) clone RP11-2503J9, which contained coding sequences for transmembrane phosphatase with tensin homology, although this gene was not disrupted. On chromosome 18p, the break point was narrowed to BAC clone RP11-527H14. In silico sequence analysis of this clone identified possible pseudo genes and gene fragments but no intact genes. RP11-527H14 also showed sites of cross hybridization, including 21p11.1. To test for a position effect on 18p11 sequences translocated to 21p11, we performed quantitative RT-PCR to measure the expression of the candidate gene C18orf1 in translocation carriers, but found no significant differences from controls in lymphoblastoid cells.
Similar content being viewed by others
Introduction
Although psychotic disorders have been shown by family and twin studies to comprise a heritable factor, it has proven difficult to isolate the genes responsible. Linkage and association studies often yield valuable but sometimes-conflicting reports for schizophrenia and bipolar disorder (grouped together as functional psychoses). The pericentric region of chromosome 18 was first shown by linkage analysis to be a susceptibility locus for bipolar disorder.1, 2 Additional linkage analyses and association studies suggested that the susceptibility could be widened to include schizophrenia,3, 4, 5 although there are non-replicated linkage reports for both diseases.3, 6, 7 These discrepancies are primarily caused by sample heterogeneity and limited statistical power in the analysis of complex diseases, although prior limited marker density is improving.
Despite this, a number of potential genes of interest have been analyzed from this area. A functional candidate, the α subunit of the olfactory G-protein Golf (GNAL) maps to the 18p locus and showed linkage and association to functional psychoses,8 although a study by Zill et al.9 could find no association between GNAL and major depression. Another functional and positional candidate, the myo-inositol monophosphatase 2 gene (IMPA2) showed evidence of association with schizophrenia and bipolar disorder in Japanese samples.5, 10 An 18p11 locus encompassing the gene C18orf1, also shows association with schizophrenia in Japanese samples.11 This gene is of unknown function but protein motifs suggest that it may interact with signaling molecules.12
Additional gene mapping information can be obtained from the identification of chromosomal translocations associated with psychiatric diseases and where possible, combining karyotyping with genetic mapping could facilitate the identification of susceptibility genes in psychiatric disorders. MacIntyre et al.13 have compiled a list of translocations co-existing with mental illnesses and tried to assess their relevance to disease based on three criteria. These are, the rarity of the translocation and independent reports of its co-existence with psychiatric illness, co-localization of the abnormality with suggestive linkage findings and co-segregation of the abnormality with illness within a family. Analysis of a chromosomal aberration that fulfilled all three criteria, a t(1;11)(q42.2;q14.3) translocation identified in a Scottish family suffering from major psychiatric illnesses, led to the isolation of genes Disrupted in schizophrenia 1 and 2 (DISC1 and DISC2).14
Smith et al.15 first identified a family whose members all carried a t(18;21)(p11.1;p11.1) translocation and suffered from either schizophrenia or related psychosis. This translocation conforms to the basic requirements set out by MacIntyre et al.,13 as being of interest to the mapping of psychiatric diseases. Here, we set out to perform a molecular analysis of the break points on both chromosomes 18 and 21.
Materials and methods
Subjects and cytogenetic analysis
All subjects examined here gave informed written consent and diagnoses were made using the DSM-IV criteria.16 Immortalized cell lines were generated from the lymphocytes of four karyotyped family members (Figure 1). High resolution GTG-banding (850 band level) was performed on the four lymphocyte cell lines derived from available family members. Twenty cells were analyzed from each cell line. We confirmed that the subjects 05 and 06 are dizygotic, by using a DNA typing kit (AmpFlSTR Profiler PCR Amplification Kit, Applied Biosystems, Foster City, CA, USA).
FISH (fluorescence in situ hybridization) analysis
For probes, BAC (bacterial artificial chromosome) clones derived from chromosome 21 were selected from the published chromosome 21 contig17 and personal communications from Dr Yaspo. BAC clone RP11-2503J9 was obtained from the Max-Planck-Institut fur Molekulare Genetik. BAC and P1 clones from chromosome 18 were selected from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) or from a physical contig map of chromosome 18p constructed by us.18 Commercially obtained BACs were purchased from Invitrogen (Carlsbad, CA, USA).
Quantitative RT-PCR
Quantitative RT-PCR was performed on mRNA from C18orf1 isolated from balanced translocation carriers (02, 04, 07), unbalanced translocation holders (06) and six controls with no translocation. The experiment was performed using the standard curve method on an ABI Prism 7900 Sequence Detection System (Applied Biosystems) and the data was captured using SDS v2.2 software (Applied Biosystems). The following oligonucleotides were synthesized from exons 5 and 6 of C18orf1:-12 C18orf1 reverse primer 5′-AAGGACGGCGCTGAAC, C18orf1 forward primer 5′-TGCCTGTGGCCTTCAGACA and C18orf1 probe FAM-5′-CCTCGGAGATCATG-TAMRA. Briefly, total RNA was extracted from the four patient cell lines and six cell lines derived from subjects free from functional psychoses and with no detectable translocation. One microgram of total RNA was used in the assay as described by the manufacturers protocol for cDNA synthesis and amplification (SuperScript Choice System, Invitrogen). The experiment was performed in triplicate and results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control.
Results
Family structure and karyotype analysis
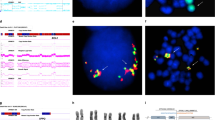
The full psychiatric history of the family has been reported earlier.15 Briefly, a female who suffered from psychosis, carried a balanced t(18;21)(p11.1;p11.1) translocation (subject 02 in Figure 1). Two sons, one of whom suffered from schizophrenia and the other from schizophrenia-spectrum disorder also carried the same balanced translocation (subjects 04 and 07 in Figure 1). A daughter with severe schizophrenia carried an unbalanced translocation, where the normal chromosome 18 was replaced by a derivative 18 (subject 06 in Figure 1). Two other siblings were unavailable for karyotypic analysis, a deceased son who suffered from psychosis and a second daughter who was reported to be well. The chromosomes 18 and 21 components of the balanced and unbalanced translocations are shown in Figures 2a and b, respectively.
Karyotype analysis of high resolution GTG-banded chromosomes showed a reciprocal translocation between band p11.1 of chromosomes 18 and 21. Three patients (02, 04 and 07 in Figures 1 and 2a) carried a balanced translocation, whereas in one patient (06 in Figures 1 and 2b), the normal chromosome 18 was replaced by the derivative 18 from the t(18;21)(p11.1;p11.1). This translocation was the sole detectable anomaly. This supported the original karyotype15 and confirmed that the cell line had not undergone further rearrangements.
It is suggested that the subject 06 carrying three derivative chromosomes could be severer than those of 04 and 07 who both have two translocations if the chromosomal abnormality contributes to develop mental illnesses in this family: indeed the subject 06, 43 years old at examination, has never been employed, living with her mother.15 In contrast, the subject 04 managed to graduate high school in an honors program and, despite his illness, he served in the US Navy for 4 years. At examination, he was a 46-year-old supermarket worker.15 The subject 07 was, 41 years old at examination, twice married and employed as an accountant.15
FISH analysis
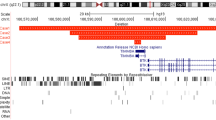
Clones were selected from contigs adjacent to or within the cytogenetic band of interest, fluorescently labeled and hybridized to metaphase spreads from control and the patient cell lines. Contiguous clones were analyzed until a split signal was observed in patient cells. We examined a total of nine clones derived from chromosome 18 and four clones from chromosome 21 by FISH (Figure 3). BAC clone RP11-2503J9 from the short arm of 21 hybridized strongly to the normal and derivative chromosome 21 and derivative chromosome 18 (Figure 4a). It also showed strong signal on multiple chromosomes, excluding chromosome 18. Signal on the derivative 18 was very weak, suggesting that the break point lay close to one end of the BAC sequence. Analysis of the BAC end sequence showed that the centromeric arm of RP11-2503J9 overlapped with the clone B15LOCO and contained partial exons for the gene transmembrane phosphatase with tensin homology (TPTE). No other genes were detected within this BAC clone. As well as B15LOCO, BAC R589M2 and P1 clones T171 and S39 all mapped to the normal and derivative chromosome 21, and therefore below the break point. The chromosome 18-derived BAC clone, RP11-527H14 showed cross hybridization to both homologs of chromosomes 18 and 21 with stronger signal on chromosome 18 in normal cells (Figure 4b). In the translocated cell line, reduced signal was seen on the derivative 18 with a concurrent increase in signal on the derivative 21 (Figure 4c). The increased signal on the derivative 21 was due to extra material translocated from chromosome 18, indicating that RP11-527H14 spanned the chromosome 18 break point.
(a) Ideogram of chromosome 21 showing markers, clone locations and chromosome of hybridization in the translocated cell line. Clone 2503J9 spans the translocation break point. (b) Ideogram of chromosome 18 showing markers, clone location and chromosome hybridization. Clone 527H14 spans the break point.
FISH mapping of BAC clones against normal and translocated cells. (a) BAC 2503J9 hybridization to the translocated cell line. The clone hybridizes to multiple chromosomes as well as the normal and derivative chromosomes 21 and the derivative 18. (b) Hybridization of 527H14 to normal cells with signal on the normal chromosomes 18 and weak signal on the normal chromosomes 21. (c) Hybridization of 527H14 to translocated cells with signal on the normal chromosomes 18 and 21, reduced signal on the derivative 18 and increased signal on the derivative 21 (*). The arrow head denotes a chromosome 21 centromeric probe, the thin arrow a chromosome 18 centromeric probe and the medium arrow, BAC signal.
In silico analysis of BAC clones
The sequence of both clones was available from the NCBI database (http://www.ncbi.nlm.nih.gov/) in draft form (AC068255 for RP11-527H14) and complete form (AF254982 for RP11-2503J9). A direct comparison of the two sequences (NCBI, pairwise Blast) produced several regions of homology. The homologous regions were predominately ALU repeats of the subfamily S and one L1 repeat. Low copy repeats (LCRs) are known to have important roles in microdeletions, chromosomal breaks, etc. According to the UCSC Human Genome Browser Gateway March 2006 assembly (http://genome.ucsc.edu/cgi-bin/hgGateway database), there are no signatures of LCRs in both the BAC sequences. Genbank contained a contig NT_024983 that included BAC RP11-527H14 sequence. The contig spanned 367 kb and encoded 16 computer-predicted exons showing homology to 4 different genes (NY-BR-1.1 is also known as ankyrin repeat domain 30B, ANKRD30B) (Table 1). However, all of the exons seemed to be derived from multicopy genes or pseudogenes.
Quantitative RT-PCR
To determine whether the translocation caused an aberrant tissue expression of the chromosome 21 derived TPTE, we performed RT-PCR on RNA derived from subject 02 and a control lymphocyte cell line with no t(18;21) translocation. No expression was detected in either the patient or control lymphocyte cell lines (data not shown). The chromosome 18 derived C18orf1 gene was also examined for a position effect based on the fact that the marker D18S1074 (Figure 3b) is located within the gene (UCSC Human Genome Browser Gateway March 2006 assembly; http://genome.ucsc.edu/cgi-bin/hgGateway), in addition to our prior association report between the gene and schizophrenia,19 but no significant differences in expression were observed between the unbalanced translocation cell line, the balanced translocation cell lines or control cell lines with no detectable translocation (Figure 5). The other candidate genes on 18p, GNAL and IMPA2 gene could not be detected by RT-PCR from lymphocyte cell line-derived RNAs.
Bar chart showing expression of C18orf1 in translocation positive and negative cell lines. Values are expressed relative to the endogenous control GAPDH (mean±s.d.). The numbers of cell lines examined were n=3 for the balanced translocation, n=1 for the unbalanced translocation, n=6 for no translocation. Triplicate experiments were done for each cell line.
Discussion
Chromosomal translocations that co-segregate with mental illness could provide a useful approach for identifying candidate genes from susceptibility loci. In this study, carriers of a balanced t(18;21)(p11.1;p11.1) translocation (02, 04, 07) suffered to varying degrees from schizophrenia or related psychosis (Figure 1). In subject 06 (Figures 1 and 2b), the normal chromosome 18 was replaced by a der18(18;21)(p11.1;p11.1), resulting in 18p11.1-18p ter sequences fused to 21p as the sole chromosome 18 complement in these cells. This patient suffered from chronic schizophrenia.
The chromosome 21 mapping and sequencing consortium generated a contig of four BAC clones and two P1 clones spanning approximately 280 kb of chromosome 21p.17 This sequence identified the gene, TPTE as the only gene on 21p.17, 20 The BAC clone 2503J9 spanned the translocation break point and overlapped with BAC, B15LOCO but was not part of the published contig. Both BACs contained exons for TPTE, making this gene closest to the break point. TPTE is expressed exclusively in the testis with multiple homologs on chromosomes 21, 13, 15, 22 and Y.20 It is not known whether the copy of TPTE on 21p is functionally active. RT-PCR experiments did not show any evidence of aberrant TPTE expression in the lymphocytes of translocation carriers suggesting that the translocation caused no functional changes on chromosome 21.
Normal primate centromeres are characterized by α satellite DNA, composed of a diverged 171 bp motif repeated in a tandem head to tail fashion and at a functional level, are transcriptionally inactive.21 The pericentric region represents a bridge between the heterochromatic centromere sequences and euchromatic DNA. The short arms of acrocentric chromosomes, such as 21 are also characterized by satellite DNAs I, II, III, IV and β.22 These regions display gene silencing through the binding of heterochromatin proteins. The translocation spanning BACs were examined for the presence of satellite repeats. BAC 2503J9 (chromosome 21) scored extensive homology with all satellite repeats. BAC 527H14 (chromosome 18) scored only one hit for an α satellite repeat. This suggested that the mechanism for the translocation differs from that of Robertsonian translocations, which are thought to be mediated by similar subtypes of satellite, repeats found in heterochromatic regions.22
An in silico search for coding sequences in BAC 527H14 identified 16 predicted exons with homology to four known genes: breast cancer antigen (ankyrin repeat domain 30B), human melanoma antigen, keratinocyte growth factor precursor and olfactory receptor pseudogene (Table 1). The lack of intron–exon boundary structure suggests that the human melanoma antigen sequence, probably represents a processed pseudogene, a common feature of pericentric regions.17 All of the putative exons map to multiple chromosomal sites, 18 and 21p or 21q as well as either chromosomes 5, 9 and 11. The lack of α satellite sequences and the presence of pseudogenes suggest that the chromosome 18 break point may lie close to the boundary between the pericentromere and transcriptionally active euchromation.
The translocation results in the fusion of genes in the region 18p11.1 to the terminus of chromosome 18 onto the short arm of chromosome 21. For the genes adjacent to the transcriptionally silent chromosome 21 sequences, a position effect could occur resulting in silencing of 18p genes. Gene silencing has been observed when heterochromatic satellite repeats are introduced close to euchromatic genes. The silencing effect appeared stable through successive cell divisions.23 None of the coding sequences from 18p11 identified in this study could be examined for position effects primarily because the exons existed as multiple copies and most were pseudogenes. We studied C18orf1 for aberrant epigenetic regulation through translocation-induced heterochromatization. As IMPA2 and GNAL are not expressed in lymphocytes, they could not be assayed by this method. Quantitative RT-PCR failed to show any significant difference in expression of C18orf1 between control cell lines and cell lines with balanced or unbalanced chromosome 18 translocations. This implies that if a position effect occurs in translocation carriers, it does not extend to C18orf1 that lies at least 2.5 Mb (assumed from the sequence of BAC 527H14 and the position of D21S318 on it) away from the break point. The same conclusion can be drawn for the genes GNAL and IMPA2 because they map to a ∼2 Mb region that includes C18orf, with C18orf1 being most proximal (UCSC Human Genome Browser Gateway March 2006 assembly; http://genome.ucsc.edu/cgi-bin/hgGateway).
The limitations of this study include: (1) we could not examine the effects of the chromosomal translocation on the candidate gene expressions in the brain, and (2) we did not examine the transcripts other than those from integral genes and all kinds of transcripts including non-coding RNAs should also be tested. Regarding the latter (2), the progress of ‘ENCODE’ (Encyclopedia of DNA Elements) project24 would be helpful to resolve the issue.
In summary, this study could provide a useful starting point for narrowing down and eventually identifying the susceptibility gene(s) on 18p, which is thought to undergo aberrant epigenetic regulation by the translocation in the current schizophrenic family.
Conflict of interest
The authors declare no conflict of interest.
References
Berrettini, W. H., Ferraro, T. N., Goldin, L. R., Weeks, D. E., Detera-Wadleigh, S., Nurnberger, Jr J. I. et al. Chromosome 18 DNA markers and manic-depressive illness: evidence for a susceptibility gene. Proc. Natl Acad. Sci. USA 91, 5918–5921 (1994).
Detera-Wadleigh, S. D., Badner, J. A., Berrettini, W. H., Yoshikawa, T., Goldin, L. R., Turner, G. et al. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc. Natl Acad. Sci. USA 96, 5604–5609 (1999).
Schwab, S. G., Hallmayer, J., Lerer, B., Albus, M., Borrmann, M., Honig, S. et al. Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am. J. Hum. Genet. 63, 1139–1152 (1998).
Van Broeckhoven, C. & Verheyen, G. Report of the chromosome 18 workshop. Am. J. Med. Genet. 88, 263–270 (1999).
Yoshikawa, T., Kikuchi, M., Saito, K., Watanabe, A., Yamada, K., Shibuya, H. et al. Evidence for association of the myo-inositol monophosphatase 2 (IMPA2) gene with schizophrenia in Japanese samples. Mol. Psychiatry 6, 202–210 (2001).
Maziade, M., Roy, M. A., Rouillard, E., Bissonnette, L., Fournier, J. P., Roy, A. et al. A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol. Psychiatry 6, 684–693 (2001).
DeLisi, L. E., Shaw, S. H., Crow, T. J., Shields, G., Smith, A. B., Larach, V. W. et al. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am. J. Psychiatry 159, 803–812 (2002).
Tsiouris, S. J., Breschel, T. S., Xu, J., McInnis, M. G. & McMahon, F. J. Linkage disequilibrium analysis of G-olf alpha (GNAL) in bipolar affective disorder. Am. J. Med. Genet. 67, 491–494 (1996).
Zill, P., Engel, R., Baghai, T. C., Zwanzger, P., Schule, C., Minov, C. et al. Analysis of polymorphisms in the olfactory G-protein Golf in major depression. Psychiatr. Genet. 12, 17–22 (2002).
Ohnishi, T., Yamada, K., Ohba, H., Iwayama, Y., Toyota, T., Hattori, E. et al. A promoter haplotype of the inositol monophosphatase 2 gene (IMPA2) at 18p11.2 confers a possible risk for bipolar disorder by enhancing transcription. Neuropsychopharmacology 32, 1727–1737 (2007).
Kikuchi, M., Yamada, K., Toyota, T., Itokawa, M., Hattori, E., Yoshitsugu, K. et al. Two step association analyses of the chromosome 18p11.2 region in schizophrenia detect a locus encompassing c18orf1. Mol. Psychiatry 8, 467–469 (2003).
Yoshikawa, T., Sanders, A. R., Esterling, L. E. & Detera-Wadleigh, S. D. Multiple transcriptional variants and RNA editing in C18orf1, a novel gene with LDLRA and transmembrane domains on 18p11.2. Genomics 47, 246–257 (1998).
MacIntyre, D. J., Blackwood, D. H., Porteous, D. J., Pickard, B. S., Muir, W. J. Chromosomal abnormalities and mental illness. Mol. Psychiatry 8, 275–287 (2003).
Millar, J. K., Wilson-Annan, J. C., Anderson, S., Christie, S., Taylor, M. S. & Semple, C. A. et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423 (2000).
Smith, A. B., Peterson, P., Wieland, J., Moriarty, T. & DeLisi, L. E. Chromosome 18 translocation (18;21) (p11.1;p11.1) associated with psychosis in one family. Am. J. Med. Genet. 67, 560–563 (1996).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association, Washington, DC, 1994).
Hattori, M., Fujiyama, A., Taylor, T. D., Watanabe, H., Yada, T., Park, H. S. et al. The DNA sequence of human chromosome 21. Nature 405, 311–319 (2000).
Reyes, G. D., Esterling, L. E., Corona, W., Ferraren, D., Rollins, D. Y. & Padigaru, M. et al. Map of candidate genes and STSs on 18p11.2, a bipolar disorder and schizophrenia susceptibility region. Mol. Psychiatry 7, 337–339 (2002).
Kikuchi, M., Yamada, K., Toyota, T., Itokawa, M., Hattori, E., Yoshitsugu, K. et al. Two-step association analyses of the chromosome 18p11.2 region in schizophrenia detect a locus encompassing C18orf1. Mol. Psychiatry 8, 467–469 (2003).
Chen, H., Rossier, C., Morris, M. A., Scott, H. S., Gos, A., Bairoch, A. et al. A testis-specific gene, TPTE, encodes a putative transmembrane tyrosine phosphatase and maps to the pericentromeric region of human chromosomes 21 and 13, and to chromosomes 15, 22, and Y. Hum. Genet. 105, 399–409 (1999).
Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K. & Willard, H. F. Genomic and genetic definition of a functional human centromere. Science 294, 109–115 (2001).
Bandyopadhyay, R., Berend, S. A., Page, S. L., Choo, K. H. & Shaffer, L. G. Satellite III sequences on 14p and their relevance to Robertsonian translocation formation. Chromosome Res. 9, 235–242 (2001).
Tulin, A. V., Naumova, N. M., Aravin, A. A. & Gvozdev, V. A. Repeated, protein-encoding heterochromatic genes cause inactivation of a juxtaposed euchromatic gene. FEBS Lett. 425, 513–516 (1998).
ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Acknowledgements
We acknowledge the assistance of Dr ML Yaspo in providing the BAC clone RP11-527H14. This work was supported by RIKEN BSI Funds, Grant-in-aid for scientific studies from the MEXT of Japan to TY and YI and CREST funds from the Japan Science and Technology Agency, Japan to TY.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meerabux, J., Ohba, H., Iwayama, Y. et al. Analysis of a t(18;21)(p11.1;p11.1) translocation in a family with schizophrenia. J Hum Genet 54, 386–391 (2009). https://doi.org/10.1038/jhg.2009.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.47
Keywords
This article is cited by
-
Two novel genomic regions associated with fearfulness in dogs overlap human neuropsychiatric loci
Translational Psychiatry (2019)
-
Alzheimer’s disease-associated (hydroxy)methylomic changes in the brain and blood
Clinical Epigenetics (2019)