Abstract
The Liqian people in north China are well known because of the controversial hypothesis of an ancient Roman mercenary origin. To test this hypothesis, 227 male individuals representing four Chinese populations were analyzed at 12 short tandem repeat (STR) loci and 12 single nucleotide polymorphisms (SNP). At the haplogroup levels, 77% Liqian Y chromosomes were restricted to East Asia. Principal component (PC) and multidimensional scaling (MDS) analysis suggests that the Liqians are closely related to Chinese populations, especially Han Chinese populations, whereas they greatly deviate from Central Asian and Western Eurasian populations. Further phylogenetic and admixture analysis confirmed that the Han Chinese contributed greatly to the Liqian gene pool. The Liqian and the Yugur people, regarded as kindred populations with common origins, present an underlying genetic difference in a median-joining network. Overall, a Roman mercenary origin could not be accepted as true according to paternal genetic variation, and the current Liqian population is more likely to be a subgroup of the Chinese majority Han.
Similar content being viewed by others
Introduction
The Liqian people live in Yongchang County (2,000 m above sea level) in the Gansu Province, China. A small proportion of the Liqian people have been described as having mixed racial morphological traits. In recent years, the Liqian population has become well known because the people were regarded as descendents of ancient Roman legionnaires, who disappeared from European history after the battle of Carrhae in southeastern Turkey in 53 B.C.E. (Dubs 1955). Homer Hasenphlug Dubs speculated that these survivors made their way east to Uzbekistan to enlist with Jzh Jzh (Dubs 1957). After the Jzh Jzh war in 36 B.C.E., more than 100 Jzh Jzh soldiers, described as Raman soldiers, settled in the present Yongchang County of Gansu province in China (Harris 1992; Song et al. 2005). To verify this hypothesis, more and more scholars have focused on studying the origin of the Liqians (Harris 1992; Dubs 1957; Song et al. 2005). Due to the lack of historical and archeological evidence, this hypothesis, based on speculation and imagination, remains controversial. Without any other direct evidence, genetic contribution seems particularly necessary. Single nucleotide polymorphisms (SNP) and short tandem repeat (STR) loci on the non-recombining region of the human Y chromosome (NRY) have been widely used to trace the origin and migratory events of modern paternal genetic lineages (Jobling and Tyler-Smith 2003). Therefore, Y-chromosome polymorphisms in our study were used to investigate the paternal genetic landscape of the Liqians and to provide genetic evidence for a suggested origin of the Liqian people.
Materials and methods
Sample collection and DNA extraction
Blood samples were obtained from 227 unrelated male individuals representing four populations from northwest China: A total of 39 Tibetans from Guide County of Qinghai Province, 49 Uygurs from Urumqi city of Xinjiang Uygur Autonomous, 87 Liqian individuals from Yongchang County of Gansu province, and 52 Yugurs from Su’nan Yugur Autonomous County of Gansu province (Fig. 1). DNA was extracted from fresh blood by use of standard phenol–chloroform protocols.
Typing of Y chromosome DNA polymorphisms
We typed 12 Y-SNPs by size detection, restriction fragment length polymorphism (RFLP) and denaturing high performance liquid chromatography (DHPLC). There were three ins/del polymorphisms detected by agarose electrophoresis, YAP (Hammer 1994), 12f2 (Rosser et al. 2000) and MSY2 (Bao et al. 2000). Some of the markers were typed by RFLP, including SRY10831 (Whitfield et al. 1995), M9, M17 (Underhill et al. 1997), Tat (Zerjal et al. 1997) and RPS4Y (Bergen et al. 1999), and the others were genotyped by denaturing high performance liquid chromatography (DHPLC), including M122, M175, M45 and M89 (Underhill et al. 2001b).
The 12 Y-chromosomal short tandem repeats (Y-STRs) are included in the Promega Corporation PowerPlex® Y System (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a, DYS385b, DYS437, DYS438 and DYS439) and were typed in 227 individuals of our study. Allele scoring for each marker was obtained by comparison to the PowerPlex® Y System allelic ladder (Promega Corporation, Madison, WI). An Applied Biosystems 3100 automated sequencer in combination with GenescanTM 3.1 analysis software was used to detect and size the amplified fragments.
Data analysis
Arlequin2.0 software (Schneider et al. 2000) was used to compute Rst distances and associated P values based on 10,000 permutations (Slatkin 1995), to test the correlation between genetic and geographic distances (distances based on the geographical center of each region) (Mantel 1967) and to measure haplogroup/haplotype diversity (Nei 1987). Principal component analysis based on Y-SNP haplogroup frequencies and multidimensional scaling analysis based on Rst matrix were carried out using the SPSS12.0. Neighbor-joining tree on the basis of microsatellite allele frequencies was obtained using programs in the Phylip (v. 3.5) package (Felsenstein 2001). Program Adimix2.0 (Dupanloup and Bertorelle 2001) was used to calculate the admixture proportions based on Y-STR data. Median-joining network analysis (Bandelt et al. 1999) of haplogroup-associated haplotypes was performed by using NETWORK4.0.
Results
Haplogroup/ haplotype distributions and associated diversity
All 12 binary markers were found to be polymorphic, and these defined 11 haplogroups (Fig. 2). The diversity of the Yugur, the Han Chinese and the Uygur populations were high; in contrast, the Liqians presented low genetic diversity with a single highest frequency haplogroup O3-M122 (71.3%) (Fig. 2). The haplogroup O3 showed moderate frequency in the Yugurs, the Inner Mongolian and the Han Chinese populations (Xue et al. 2006) and low frequency in the Tibetans and the Uygurs. In our study, haplogroup DE defined by YAP occurred at low haplogroup frequency in Liqians, and it was also prevalent in the Yugurs, but amounted to 48.7% in the Tibetans. The lineage characterized by RPS4Y is predominant in the Inner Mongolians (Xue et al. 2006), with moderate frequency in Han Chinese and Yugur populations, but it is rare in Liqian and other Chinese populations (Xue et al. 2006). R1a1*-M17, prevalent in the Middle East and Europe (Luis et al. 2004), amounts to 28.6% in the Uygurs, while it was rare in most Chinese populations, including the Liqian and Yugur people. Obviously, the lineages (I, J and H), frequent in West Eurasia (Semino et al. 2000), are lacking in Liqian people, whereas the haplogroup O, prevalent in Liqian and other East Asian populations, is lacking in Central Asia and West Eurasia.
Further analysis performed on Y-STR outlined a more detailed view of Liqian Y chromosome variation. The 227 Y chromosomes representing the four populations were typed with 12 Y-specific microsatellites. Two hundred nine complete different haplotypes were identified, among which 75 haplotypes belong to Liqian people. Haplotype diversities were higher than 0.98.
Worldwide data for the 12-element haplotype are unavailable in the Y-STR Haplotype Reference Database (YHRD) (Roewer et al. 2001), but a subset of 9 of the STRs is included. A comparison on the basis of the minimal haplotype yielded 16 matches, most of which (12/16) belong to East Asia and South Asia. For the 4 of 16 matches, 2 are found only in African populations, and the other 2 are shared by European and Asian populations. All four Liqian Y chromosomes belong to haplogroup O3-M122. No West Eurasian-specific matches were observed in Liqian Y chromosomes.
Principal component analysis
For comparative analysis, principal component analysis of Y haplogroup frequencies included published data from Chinese, Outer Mongolian, Central Asian and West Eurasian populations (Fig. 3). The PC plot based on haplogroups shared by all populations reveals that the Liqians are close to most populations in China and clearly distinguished from the Central Asian and West Eurasian populations. Particularly, the Liqians are closely related to the Yugur, Han Chinese and Mongolian populations.
Principal component analysis based on Y-SNP haplogroup frequencies in China and Outer Mongolia (Xue et al. 2006), Central Asia (Wells et al. 2001) and West Eurasia (Wells et al. 2001; Semino et al. 2000). The proportion of the variation explained by PC1 is 61.15%, which explained by PC2 is 17.83%. Squares represent Central Asian, West Eurasian and Outer Mongolian populations, and triangles represent Chinese populations
Rst values and MDS analysis
Microsatellite allele frequency data were used to calculate pairwise Rst genetic distances and to perform MDS analysis. Due to the lack of original STR data of populations from Central Asia, pairwise Rst values based on nine STRs (minimal haplotype) and subsequent MDS analysis were restricted to China, West Eurasia and Outer Mongolia (Fig. 4). Most Y-STR data taken from the literature were collected by using high-resolution genotyping technologies, which made further genetic analysis more reliable. The Liqian population shows non-significant Rst values compared to the Yugur and Han Chinese populations mentioned in our study and shows significant Rst values to all other populations (data not shown). Fig. 4 portrays the results of MDS. When all populations were included, these populations lying around the outside, with extreme pairwise Rst values, cause a compression of all the remaining populations in the MDS plot. Thus, most populations cluster in the center. To extend this observation, we reanalyzed the central group of populations, which were not marked in Fig. 4a. A good fit between the two-dimensional plot and the source data (pairwise values of Rst) was obtained in Fig. 4b, demonstrated by the low stress value (0.037). The Liqian and the Han Chinese populations were placed in the lower left part of the plot, somewhat distinct from the Mongolians and Yugurs, and quite separate from the West Eurasian populations.
Two-dimensional MDS plot based on pairwise Rst values in populations throughout Eurasia and b populations lying in the center and not marked in Fig. 4a. Populations from West Asia [Arab (Immel et al. 2004), Anatolian (Çakır et al. 2004)], Europe [Albanian (Robino et al. 2002), North Italian (Cerri et al. 2005), Croatian (Lovrečic et al. 2005), Sicilian (Robino et al. 2005), Greek (Robino et al. 2004), Central Italy (Ricci et al. 2001), Gypsy and Macedonian (Peričić et al. 2005) and China (Han(He’nan) (Feng et al. 2005), Han (Liaoning) (Wang et al. 2006), Han (Ningxia)(Zhu et al. 2006c), Mongolians (Zhu et al. 2005a), Tibetans (Zhu et al. 2006b), Uygurs(Yili) (Zhu et al. 2005b), Tibetans (Qinghai), Uygurs(Urumqi), Yugurs and Liqians (present study)]
Mantel test
Microsatellite loci with high mutation rates are more suitable to study recent human evolution than binary markers (Karafet et al. 2001). We therefore based the subsequent analysis on Y-STR data. To test the correlation between genetic and geographic distances, the Mantel test was performed for Liqian, Mongolian, Yugur and Han Chinese populations, which are closely related to the Liqian people in MDS plots (Fig. 4). The Mantel test is based on pairwise Rst values computed by using 11 Y-STRs because the DYS 437 was not analyzed in Mongolian and Han Chinese populations in Liaoning and Ningxia. The result showed a significantly positive correlation between pairwise genetic distances (Rst matrix) and pairwise geographic distances (r = 0.646, P = 0.003) (Table 1), indicating that genetic distances are linearly related to geographical distances, as observed in northern groups of East Asia (Karafet et al. 2001).
Phylogenetic analysis
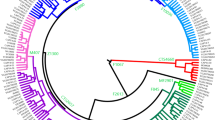
In MDS analysis, the Liqian population is closely related genetically to the Han Chinese, Inner Mongolian and Yugur populations. In order to better understand their relationship, we constructed a neighbor-joining phylogenetic tree. Tunisian (Frigi et al. 2006) was considered as an outgroup. Thus, the phylogenetic analysis included the Han Chinese, Yugur, Tunisian and Inner Mongolian populations in Fig. 4. Therefore, phylogenetic analysis was performed on only 11 STR markers (not include DYS437) shared by the 7 populations in Fig. 5. The relationships among these populations were illustrated in the tree (Fig. 5). The Liqian and Han Chinese populations in He’nan and Ningxia are scattered on one branch. The Mongolians, Han Chinese in Liaoning and Yugur formed a separate branch, respectively. In all, the Liqian people are closely related to adjacent Han Chinese populations, and slightly distinguished from the Yugur in Fig. 5.
Admixture analysis
PC and MDS analysis suggests that the Liqians are close to Chinese populations. If we assume that Liqians are a genetic admixture of some parental populations, then the Han Chinese and Mongolian populations should be representatives of these populations. Although the Yugurs are genetically close to the Liqian, they were excluded in the admixture analysis because the Yugur people were considered a kindred population (Song et al. 2005), and the Yugur people are younger than the Liqian population (Song et al. 2005; Hao 1999). The mY estimate is based on a coalescent approach and is suitable for analyzing microsatellite data (Bertorelle and Excoffier 1998). Therefore, for the same reasons as for the Mantel test, the present admixture analysis is based only on the data of 11 Y-STRs. Our results suggest that a large majority of Liqian Y chromosomes has a Han Chinese origin and that a small Mongolian contribution was observed in the Liqians (Table 2). Simultaneously, by using the same program, the admixture estimates were performed on the Yugur people in order to test the relationship of the Liqian and Yugur people. The result indicated that the Liqian people and the Yugurs have similar proportional contributions from the Han Chinese and Mongols, which is compatible with the historical hypothesis that the Yugurs and Liqians are kindred populations with common origins (Song et al. 2005).
Median-joining network
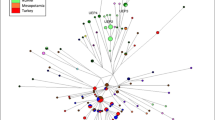
Principal component and admixture analyses suggest that the Liqians are closely related to the Yugurs; in contrast, an underlying difference between the Liqian and Yugur people has been observed in phylogenetic analysis (Fig. 5). To further understand the relationship of the Liqian and Yugur populations, a median-joining network of haplotypes associated with haplogroup O was constructed (Fig. 6). The network grouped these Y chromosomes into two clusters. In the network, Liqian chromosomes form two clusters, one almost distinct and the other shared with that involving most Yugur Y chromosomes. It is interesting to note that the Yugur cluster is surrounded by Liqian Y chromosomes. In general, mutation, recombination and migration can increase variation (Zerjal et al. 2003). According to this, the Liqian people may have a more complex demographic history than the Yugur people. The feature explains the close genetic affinity accompanied by an underlying genetic difference between Liqian and Yugur populations and also reflects that the haplotype diversity of Liqians is higher than that of the Yugurs within haplogroup O. Overall, the Liqian and the Yugur haplotype associated with haplogroup O are completely intermingled.
Median-joining network of haplotypes based on 12 Y-STRs associated with haplogroup O of Liqian and Yugur chromosomes. Circles have an area proportional to haplotype frequency. The smallest circle of the network corresponds to a single Y chromosome. Liqian Y-chromosomes are represented in gray, Yugurs in black
Discussion
The first extensive analysis of Y variation in the Liqians was carried out in our study. This allowed us to compare our data with those previously reported in worldwide populations in order to investigate the origin and evolution of the Liqian people. The results based on Y-SNPs are consistent with those obtained with Y-STRs, which reinforce our findings.
In the present study, it was unexpected that 71.3% Y chromosomes of the Liqians belonged to the haplogroup O3-M122, which is an East Asian-specific haplogroup (Su et al. 1999; Shi et al. 2005). The O3-M122 frequency in the Liqians was by far the highest one observed among populations in northern China (Xue et al. 2006; Su et al. 1999, 2000b; Shi et al. 2005). Since the Han dynasty, the Liqians have been living in northwestern China, which has been well-established by historical records (Song et al. 2005). Combining with the historical records, the haplogroup distribution suggested that most of the Liqian Y chromosomes could be traced back to northern China. The result was also reflected in PC and MDS analysis, which indicated that the Liqians were genetically close to Chinese populations, especially Han Chinese populations in various regions. The result was confirmed in admixture and phylogenetic analysis (Table 2, Fig. 5), suggesting a strong Han Chinese paternal influence on the Liqian gene pool. The Mantel test suggests that the Liqian people, wherever they originated, must have had an extensive gene exchange with the local people. Besides the Han Chinese people, Mongolians and Yugurs in China are genetically related to the Liqian people.
Previous studies demonstrated that the male lineages of the Mongolians spread rapidly in a large part of Asia (Zerjal et al. 2002, 2003). The Hexi region in northern China, where the Liqians have settled, was controlled by the Mongols in the 13th and 14th centuries. It is reasonable that the Mongolians would have an impact on the Liqian gene pool. In our study, a small Mongolian contribution was observed in the Liqian gene pool.
The Yugurs, described as a kindred population of the Liqians, were genetically close to the Liqians in PC and MDS plots, which is compatible with the admixture analysis. The paternal genetic contribution indicated that the Liqian and the Yugur populations have similar contribution proportions from the Han Chinese and Mongolian populations. Nevertheless, the underlying genetic difference between the Liqian and Yugur populations was explored in a median-joining network and neighbor-joining tree (Fig. 5, Fig. 6). It may result from long-term isolation by distance. In addition, the Yugurs are strictly endogamous and live in autonomous regions of the central province of Gansu, while the Liqians live together with Han Chinese. Overall, the genetic difference between the Liqian and Yugur populations is statistically non-significant.
In PC and MDS analysis, the Liqian population is fairly distinguished from Central Asian and West Eurasian populations. The result is incompatible with the historical hypothesis that the Liqian people derived from ancient Roman soldiers, which probably included mercenaries from West Asia, as described by Huang et al. 1990. When Liqian haplotypes were compared with worldwide populations in the YHRD, no matches were found only in the West Eurasian populations, and only two matches were shared by European populations and East Asian populations. The two Liqian haplotypes are present in West European populations, but they belong to East Asian-specific haplogroup O3. The incompatible fact probably arose from the high mutation rate of Y-STR. It seemed that the two Y chromosomes are more likely to be Asian lineages than European lineages. Failure to find an apparent link between the Liqian people and ancient Roman soldiers in this study might be either because long distance migration and intermarriage have erased earlier genetic signatures or because the Liqians are just a general population in north China.
Moreover, it is noteworthy that a small proportion of Liqian people with mixed racial traits are not necessarily associated with ancient Roman soldiers. Along the ancient Silk Road in north China, it is common to see people with Caucasian morphological traits, which is also a classical trait of Chinese minority ethnic groups in Xinjiang (like the Uygurs). Therefore, we cannot trace a Liqian origin only from morphological traits.
As described above, the Liqians are closely related to Chinese populations, especially the Han Chinese in north China. In addition, the Liqian and Yugur populations are likely to be kindred populations. No obvious signature of Roman soldier origin is observed in the Liqian paternal gene pool. A Roman mercenary origin for the Liqian people is likely to be nothing more than an interesting theory. In order to reveal genetic landscapes of the Liqians completely, complementary autosomal and mtDNA studies have to be carried out in future work.
References
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bao W, Zhu S, Pandya A, Zerjal T, Xu J, Shu Q, Du R, Yang H, Tyler-Smith C (2000) MYS2: a slowly evolving minisatellite on the human Y chromosome which provides a useful polymorphic marker in Chinese populations. Gene 244:29–33
Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D (1999) An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80
Bertorelle G, Excoffier L (1998) Inferring admixture proportions from molecular data. Mol Biol Evol 15:1298–1311
Çakır AH, Çelebioğlu A, Yardımcı E (2004) Y-STR haplotypes in Central Anatolia region of Turkey. Forensic Sci Int 144(1):59–64
Cerri N, Verzeletti A, Bandera B, De Ferrari F (2005) Population data for 12 Y-chromosome STRs in a sample from Brescia (northern Italy). Forensic Sci Int 152:83–87
Dubs HH (1957) A Roman city in ancient China. The China Society, London
Dupanloup I, Bertorelle G (2001) Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol 18(4):672–675
Feng CJ, Xiang ZD, Shen CB (2005) Polymorphisms of twelves Y-chromosome STR loci in Han population in Henan. Penal Technic(China) 3:23–28
Felsenstein J (2001) PHYLIP: phylogeny inference package. University of Washington, Seattle
Frigi S, Pereira F, Pereira L, Yacoubi B, Gusmao L, Alves C, Khodjet el Khil H, Cherni L, Amorim A, El Gaaied A (2006) Data for Y-chromosome haplotypes defined by 17 STRs (AmpFLSTR1 YfilerTM) in two Tunisian Berber communities. Forensic Sci Int 160:80–83
Hammer MF (1994) A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11:749–761
Hao SM (1999) On the cultural characteristics of special ethnic groups in Gansu and Qinhai. Ethnic Press, China
Harris D (eds) (1992) Black horse odyssey: search for the lost city of Rome in China. Wakefield Press, South Australia
Huang HX, Lu YT, Wu PP (translator) (1990) The life of the noble Grecians and Romans. The Commercial Press, Beijing, China
Immel UD, Kleiber M, Klintschar M (2004) Y-chromosomal STR haplotypes in an Arab population from Yemen. Int Congress Series 1261:340–343
Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612
Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF (2001) Paternal population history of East Asia: source, patterns, and microevolutionary processes. Am J Hum Genet 69:615–628
Lovrečić L, Ristić S, Brajenović B, Kapovic M, Peterlin B (2005) Human Y-specific STR haplotypes in the Western Croatian population sample. Forensic Sci Int 149:257–261
Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ (2004) The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 74:532–544
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Peričić M, Klarić IM, Lauc LB, Janićijević B, Đordević D, Efremovska L, Rudan P (2005) Population genetics of 8 Y chromosome STR loci in Macedonians and Macedonian Romani(Gypsy). Forensic Sci Int 154:257–261
Ricci U, Sani I, Giovannucci Uzielli ML (2001) Y-Chromosomal STR haplotype in Toscany (central Italy). Forensic Sci Int 120:210–212
Robino C, Gino S, Ricci U, Grignani P, Previdere C, Torre C (2002) Y-chromosomal STR haplotypes in an Albanian population sample. Forensic Sci Int 129:128–130
Robino C, Inturri S, Gino S, Torre C, Di Gaetano C, Crobu F, Romano V, Matullo G, Piazza A (2005) Y-chromosomal STR haplotypes in Sicily. Forensic Sci Int 159:235–240
Robino C, Varacalli S, Gino S, Chatzikyriakidou A, Kouvatsi A, Triantaphyllidis C, Di Gaetano C, Crobu F, Matullo G, Piazza A, Torre C (2004) Y-chromosomal STR haplotypes in a population sample from continental Greece, and the islands of Crete and Chios. Forensic Sci Int 145:61–64
Roewer L, Krawczak M, Willuweit S, Nagy M, Alves C, Amorim A, Anslinger K, Augustin C, Betz A, Bosch E, Caglia A, Carracedo A, Corach D, Dekairelle AF, Dobosz T, Dupuy BM, Furedi S, Gehrig C, Gusmao L, Henke J, Henke L, Hidding M, Hohoff C, Hoste B, Jobling MA, Kargel HJ, de Knijff P, Lessig R, Liebeherr E, Lorente M, Martinez-Jarreta B, Nievas P, Nowak M, Parson W, Pascali VL, Penacino G, Ploski R, Rolf B, Sala A, Schmidt U, Schmitt C, Schneider PM, Szibor R, Teifel-Greding J, Kayser M (2001) Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci Int 118:106–113
Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Corte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Golge M, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Norby S, Pandya A, Parik J, Patsalis PC, Pereira L, Peterlin B, Pielberg G, Prata MJ, Previdere C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543
Schneider S, Kueffer JM, Roessli D, Excoffier L (2000) Arlequin ver 2.000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva
Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens in extant Europeans: A Y chromosome perspective. Science 290:1155–1159
Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B (2005) Y-Chromosome evidence of southern origin of the east Asian-Specific haplogroup O3-M122. Am J Hum Genet 77:408–419
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139(1):457–462
Song GR, Gu S, Chen SN (2005) Liqian investigation. Shanxi Tour Press, Xi’an, China
Su B, Xiao CJ, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L (2000b) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590
Su B, Xiao JH, Underhill PA, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724
Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001b) The phylography of Y-chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62
Wang XL, Sawaguchi T (2006) Analysis of Y-STR loci in a population sample from Northeast China. J Forensic Sci 51:195–198
Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249
Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380
Xue YL, Zerjal T, Bao WD, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C (2006) Male demography in East Asia: a north-south contrast in human population expansion times. Genetics 172:2431–2439
Y Chromosome Consortium (YCC) (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2:339–348
Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183
Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (2002) A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet 71:466–482
Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C (2003) The genetic legacy of the Mongols. Am J Hum Genet 72:717–721
Zhu B, Deng Y, Zhang F, Wei W, Chen L, Zhao J, He Y, Tian Y, Xu Y, Yu R, Fang J, Liu Y (2006c) Genetic analysis for Y chromosome short tandem repeat haplotypes of Chinese Han population residing in the Ningxia Province of China. J Forensic Sci 51:1417–1420
Zhu B, Li X, Wang Z, He Y, Zhao J, Liu Y (2005a) Y-STRs haplotypes of Chinese Mongol ethnic group using Y-PLEX 12. Forensic Sci Int 153:260–263
Zhu B, Liu S, Ci D, Huang J, Wang Y, Chen L, Zhu J, Xu Y, Zhao Q, Li S, Liu Y (2006b) Population genetics for Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic minority group in Tibet. Forensic Sci Int 161:78–83
Zhu B, Wang Z, Yang C, Li XS, Zhu J, Yang G, Huang P, Liu Y (2005b) Y-chromosomal STR haplotypes in Chinese Uigur ethnic group. Int J Legal Med 119:306–309
Acknowledgments
We express our appreciation to the original DNA donors who made this study possible. We thank Maria-Catira Bortolini for statistical assistance. This work was supported by grant of National Natural Science Foundation of China (30571014), a grant of platform of the Genetic Resources of Chinese Populations Project from the Ministry of Science and Technology of P.R. China (2003DEA3N026) and the Program for New Century Excellent Talents in the University from the Ministry of Education of P.R. China (NCET-05-0885).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, R., An, L., Wang, X. et al. Testing the hypothesis of an ancient Roman soldier origin of the Liqian people in northwest China: a Y-chromosome perspective. J Hum Genet 52, 584–591 (2007). https://doi.org/10.1007/s10038-007-0155-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0155-0
Keywords
This article is cited by
-
Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome
Molecular Genetics and Genomics (2021)
-
Population genetic study of 34 X-Chromosome markers in 5 main ethnic groups of China
Scientific Reports (2015)
-
Mexican mestizo population sub-structure: effects on genetic and forensic statistical parameters
Molecular Biology Reports (2012)