Abstract
The opportunistic pathogen Pseudomonas aeruginosa is a frequent colonizer of the airways of patients suffering from cystic fibrosis (CF). Depending on early treatment regimens, the colonization will, with high probability, develop into chronic infections sooner or later, and it is important to establish under which conditions the switch to chronic infection takes place. In association with a recently established sinus surgery treatment program for CF patients at the Copenhagen CF Center, colonization of the paranasal sinuses with P. aeruginosa has been investigated, paralleled by sampling of sputum from the same patients. On the basis of genotyping and phenotypic characterization including transcription profiling, the diversity of the P. aeruginosa populations in the sinuses and the lower airways was investigated and compared. The observations made from several children show that the paranasal sinuses constitute an important niche for the colonizing bacteria in many patients. The paranasal sinuses often harbor distinct bacterial subpopulations, and in the early colonization phases there seems to be a migration from the sinuses to the lower airways, suggesting that independent adaptation and evolution take place in the sinuses. Importantly, before the onset of chronic lung infection, lineages with mutations conferring a large fitness benefit in CF airways such as mucA and lasR as well as small colony variants and antibiotic-resistant clones are part of the sinus populations. Thus, the paranasal sinuses potentially constitute a protected niche of adapted clones of P. aeruginosa, which can intermittently seed the lungs and pave the way for subsequent chronic lung infections.
Similar content being viewed by others
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen capable of causing chronic infections in the airways of cystic fibrosis (CF) patients, where it is the predominant pathogen associated with morbidity and mortality (Koch, 2002). Patients often acquire intermittent colonization of the lungs from early childhood, which eventually proceeds into chronic lung infection where the same bacterial lineage can persist for decades (Høiby et al, 2005; Jelsbak et al., 2007). In this process, P. aeruginosa is subjected to strong selection in the host environment leading to substantial genetic change and diversification. Many characteristics of chronically infecting strains are consistently selected in different CF patients, suggesting that adaptation occurs with conserved patterns of evolution (Nguyen and Singh, 2006; Smith et al., 2006; Yang et al., 2011). These characteristics include enhanced antibiotic resistance, changes in nutrient utilization, overproduction of the mucoid exopolysaccharide alginate, reduction in growth rate and loss of O-antigen, motility, type III secretion and quorum sensing (Lam et al., 1980; Hancock et al., 1983; Mahenthiralingam et al., 1994; Dacheux et al., 2001; D'Argenio et al., 2007; Yang et al., 2008; Hoffman et al., 2010). In addition, the initial infecting strains of P. aeruginosa often diversify into subpopulations with distinct colony morphologies (Wahba and Darrell, 1965; Thomassen et al., 1979). Major selective factors in the CF airways are believed to be the host immune system, nutrient composition and antibiotic treatments, and consequential adaptive changes of the bacterial phenotypes can seriously affect disease outcome and progression (Deretic et al., 1995; Haussler et al., 1999; Parad et al., 1999; Hoffman et al., 2009).
Initial colonizing strains primarily originate from the environment, each event usually with a unique genotype, displaying wild-type characteristics with a non-mucoid and antibiotic-susceptible phenotype (Burns et al., 2001). The early stages of lung disease have therefore been recognized as windows of opportunity to eradicate P. aeruginosa (Burns et al., 2001), and clinical trials have shown that early aggressive treatment is beneficial for the patient and delays transition into a chronic infection (Høiby et al., 2005; Taccetti et al., 2005). Following eradication, a new acquisition is often with a different genotype, but some studies have shown that in approximately 25% of the cases re-colonization occurs with the same genotype of P. aeruginosa (Munck et al., 2001; Gibson et al., 2003; Doring et al., 2006). Re-colonization could be either from a persistent environmental source or from an undetected reservoir in the patient upper airways such as the paranasal sinuses (Jelsbak et al., 2007). Only few reports have addressed the role of the upper airways, and these studies suggest that the paranasal sinuses can be a possible gateway and reservoir for lung infection in CF patients (Taylor et al., 1992; Dosanjh et al., 2000; Muhlebach et al., 2006; Mainz et al., 2009). The mechanism may be aspiration during sleep or when suffering from a viral infection (Huxley et al., 1978). The mucous membrane lining of the paranasal sinuses is similar to that of the conductive bronchi and the mucociliary clearance of the sinuses is impaired in CF patients by malfunction of the CF transmembrane regulator as is also the case in the lower airways. Therefore, the physiological properties and environment of the sinuses and lungs are similar (Aanaes et al., 2011). The consequences of bacterial colonization for the sinuses are detention of thickened mucus, chronic inflammation and sinonasal abnormalities that affect almost all patients from an early age (Robertson et al., 2008). The fair majority of CF patients acquire a chronic bacterial sinusitis with varying degrees of symptoms (Coste et al., 1995; Gysin et al., 2000; Robertson et al., 2008). When colonization of the sinuses occurs, the microbiota of the sinuses and lower airways are quite similar, and identical genotypes of P. aeruginosa (and other pathogens) can be isolated from both sites (Dosanjh et al., 2000; Muhlebach et al., 2006; Mainz et al., 2009). Little is known about the dynamics of the interchange of P. aeruginosa between upper and lower airways in the early stages of lung disease, and the role of the sinuses in the development of chronic lung infection needs further elucidation. Here we have investigated populations of P. aeruginosa in the combined airways of a number of children suffering from CF, and based on genotypic and phenotypic data, we conclude that the paranasal sinuses may constitute an important niche for bacterial adaptation from which subsequent persistent infections in the lungs of the patients may be established.
Materials and methods
Bacterial strains, plasmids and culture conditions
Strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli were routinely grown in Luria-Bertani (LB) medium or LB-agar at 37 °C, unless noted. Antibiotics were used at the following concentrations: 30 μg ml−1 gentamycin, 200 μg ml−1 carbenicillin and 100 μg ml−1 ampicillin for P. aeruginosa; and 15 μg ml−1 gentamycin and 100 μg ml−1 ampicillin for E. coli.
CF patients and clinical isolates
A total of 46 patients from the Copenhagen CF Center, Rigshospitalet, were included in a longitudinal study where first and subsequent lung colonizing isolates were stored from the beginning of 2005 until conclusion in July 2009 (Supplementary Table S1). This group included the majority of young CF patients who acquired their first P. aeruginosa within the study period. Patients were monitored on a monthly basis in the outpatient clinic by evaluation of their clinical status, pulmonary function and microbiology of lower airway secretions that were obtained by expectoration, endolaryngeal suction or bronchoalveolar lavage. Identification of P. aeruginosa in lower airway samples was carried out at the Department of Clinical Microbiology, Rigshospitalet, as described previously (Høiby and Frederiksen, 2000). The genotype of P. aeruginosa was determined by single-nucleotide polymorphism (SNP) typing using the AT-Biochip (Clondiag, Jena, Germany) as described by the manufacturer. Three months of oral ciprofloxacin and inhaled colistin were used to treat patients at first acquisition of P. aeruginosa as well as patients with intermittent colonization. Intravenous treatment was started if recurrent isolate during treatment, mucoid phenotype or increased antibodies occurred. Chronic P. aeruginosa lung infection was defined as the persistent presence of P. aeruginosa in the sputum for 6 consecutive months or less when persistence was combined with the presence of two or more precipitating antibodies against P. aeruginosa (Høiby, 1974, 1977).
Sample materials from the paranasal sinuses were obtained by functional endoscopic sinus surgery in the ENT Department at Rigshospitalet, Copenhagen. The location and side (left or right) of each sinus sample was noted. Identification and genotyping of P. aeruginosa from the sinus samples was performed as described for the lower airway samples.
Morphotype diversity
The morphotype diversity of P. aeruginosa in the sinus samples was assessed using Pseudomonas isolation agar (PIA; Difco, Lawrence, KS, USA) plates containing ampicillin (100 μg ml−1). Sample material was spread and plates incubated overnight (ON) at 37 °C. Colonies were screened using a Zeiss axioplan microscope equipped with a × 2.5 plan objective. The number of colonies screened from each sinus surgery ranged from several hundred to several thousand depending on the colony-forming units of the samples. Several colonies of each different morphotype were picked, clonally purified on PIA amp plates and frozen at −80 °C after growth ON in LB. The isolates were then re-cultivated on PIA amp plates and the different colony morphotypes were classified by comparing the shape of single colonies grown ON simultaneously on the same batch of plates (as the colony morphologies vary with dryness of plate, crowdedness and other factors). Morphotypes were distinguished based on features such as colony size, color (dark or light), translucent or opaque, surface roughness, surface shape (mountain or flat), line pattern on top, sharp edge or fuzzy edge and form/shape of colony (round, oval and other). As only negligible intra-morphotype differences were present in the tested phenotypes, a representative of each colony morphotype was chosen for further investigations. Morphotype diversity of P. aeruginosa in the longitudinal lower airway samples were assessed as described for the sinus samples with a small modification: after spreading the sample on PIA amp plates, 96 colonies were picked (if possible) and frozen in a microtiter plate at −80 °C. Isolates were re-cultivated to asses morphotype diversity and a representative of each colony morphotype was chosen for further investigations. All unique morphotypes from one patient was assigned a capital letter, which does not correlate between patients, except morphotype M (mucoid) and SCV (small colony variant). All colony morphotypes isolated from the sinuses and lower airways of patient B11, B13, B34, B22, B28 and B42 are listed in Supplementary Table S2.
Sequencing of mucA and lasR genes
A 687-bp fragment covering the mucA region and an 1300-bp fragment covering the lasR region plus 495 bp upstream were amplified by standard polymerase chain reaction. Primers used for polymerase chain reaction amplification and sequencing of mucA were mucA1fwd (CTCTGCAGCCTTTGTTGCGAGAAG), mucA1rev (CTGCCAAGCAAAAGCAACAGGGAGG), and for lasR were lasRfwd3 (CTGGAAAAGTGGCTATGTCG) and lasRrev3 (TGCCCTTCCCTATATATCTGC).
Motility assays
All motility assays were performed using (soft) agar plates with ABT minimal medium (Hentzer et al., 2002) containing 0.5% glucose and 0.5% casamino acids. Plates were inoculated from single colonies using a sterile toothpick and incubated at 37 °C. Swimming and swarming motility was assayed on 0.3% and 0.6% agar plates (wt vol−1), respectively, incubated for 24 h and twitching motility was assayed on 1.3% agar plates (wt vol−1) incubated for 48 h. Maximum diameter of the motility zone was recorded for minimum three replicates per strain.
Quorum sensing
Clinical strains were examined for a metallic iridescent screen indicative of the presence of lasR mutations as described previously (D'Argenio et al., 2007). Confirmation of a lasR mutation was carried out by polymerase chain reaction and sequencing. Isolates were tested for the presence of C4-HSL signal molecules using pMHRA containing an RlhR-regulated rhlA::gfp(ASV) translational fusion (Yang et al., 2009). 3-Oxo-C12-HSL signal molecules were assayed using pMHLAS containing the lasB::gfp(ASV)—Plac::lasR monitor cassette (Hentzer et al., 2002). A lasI, rhlI double mutant of P. aeruginosa (JP2) containing C4 or C12 monitor plasmid was cross-streaked on LB-agar against a clinical isolate in a T-shaped pattern as described previously (Andersen et al., 2001). The plate was incubated for 24 h before it was examined for green fluorescent protein fluorescence using an Axioplan Epifluorescence Microscope (Carl Zeiss, Copenhagen, Denmark).
Protease assay
Secreted protease production was assayed using the method described in Brown and Foster (1970). Single colonies of each strain were patched onto LB-agar containing 10% skimmed milk and incubated at 37 °C for 24 h and 48 h. The diameter of the clearing zones surrounding bacterial growth was measured in triplicate experiments.
Biofilm formation
Biofilm formation was examined in 96-well U-bottom polystyrene plates using crystal violet staining (Pratt and Kolter, 1998). Briefly, ON cultures of P. aeruginosa were diluted 20 times in ABT minimal medium (Hentzer et al., 2002) containing 0.5% glucose and 0.5% casamino acids. A measure of 150 μl of diluted bacterial culture was incubated in the microtiter plates for 24 h at 37 °C. Staining was performed using a 0.02% crystal violet solution for 20 min and optical density measured at 595 nm. Results are representative of two separate experiments with a minimum of seven replicates in total for each strain.
DNA microarray sample processing
Transcriptomic profiles of clinical isolates were obtained using the Affymetrix P. aeruginosa gene chip. Cells were grown in beef broth (State Serum Institute, Copenhagen, Denmark) at 37 °C with shaking at 170 r.p.m. Triplicate experiments were performed for each strain. A 50 ml volume of the medium was inoculated with cells from an ON culture to yield a start optical density of approx. 0.1 at 600 nm. A measure of 4 ml cells were harvested at an optical density of approx. 1 at 600 nm. RNA isolation and purification was performed by RNA Protect Bacteria Reagent and RNeasy Mini Kit (Qiagen, Hilden, Germany). RQ1 RNAase-free DNAse (Promega, Madison, WI, USA) was added to remove the contaminating DNA. Processing of the P. aeruginosa GeneChip was performed at the Department of Clinical Biochemistry, Microarray Core Unit, Rigshospitalet. In all, 10 mg of purified RNA was used to synthesize single-stranded cDNA with SuperScript Choice system (Invitrogen, Taastrup, Denmark) with a random primer. DNAse I (Amersham Biosciences, Uppsala, Sweden) was used for fragmentation of the DNA followed by biotin labeling (GeneChip DNA Labeling Reagent; Affymetrix Inc., Santa Clara, CA, USA) The labeled cDNA was then hybridized on Affymetrix P. aeruginosa gene chips and stained on the GeneChip Fluidics Station. The probe arrays were scanned with the GeneChip Scanner 3000. The Fluidics Station and GeneArray Scanner were operated and managed with the GeneChip Operation Software v.1.4.
Microarray data analysis
Microarray data analysis was performed using bioconductor for the R software (http://www.bioconductor.org). Normalization and expression index calculation was carried out with the rma function. A P-value <0.05 and absolute fold change ⩾2 was applied as cutoff values. The fold changes of expression between two strains were calculated as the ratio of the average expression levels (of the three replicates). The annotations and functional classes were assigned according to the Pseudomonas Genome Database v.2 (http://www.pseudomonas.com).
Antibiotic resistance
Minimum inhibitory concentrations (MICs) were estimated by E-test according to the manufacturer's guidelines (AB Biodisk, Solna, Sweden) with minor modifications. ON cultures in LB were diluted with LB to an optical density of 0.5 at 600 nm. A measure of 100 μl was spread with a Drigalski spatula on a predried LB plate. Subsequently E-test strips were carefully placed on the LB plate and MIC values were read after 24 h of incubation at 37 °C. A minimum of two replicates were performed for each strain (if results were not consistent more replicates were performed).
Results
In children suffering from CF, it was previously found that early airway infections with P. aeruginosa can be effectively removed from the lungs by antibiotic treatment, but often the same clones reappeared after months of clearance and it was suggested that subpopulations of the bacteria were hiding in the sinuses (Jelsbak et al., 2007). The primary hypothesis of this study therefore has been that the paranasal sinuses of CF children constitute a protected environment in which P. aeruginosa may hide to avoid the antibiotics and the immune system of the host, and from this niche subsequent seeding of the bacteria to the lower airways can take place. The hypothesis predicts that children with recurrent intermittent colonization of the lower airways with the same genotype of P. aeruginosa would also be expected to have their sinuses colonized with the same genotype. This hypothesis was tested in several stages: First, by genotyping isolates from the different airway compartments to establish clonal relationships, second to assess the population structure of P. aeruginosa in the airways, third to characterize the developed phenotypes in the different airway populations and finally to find evidence for the direction of migration of the evolved lineages between the airway compartments.
Airway colonization and P. aeruginosa genotyping in intermittently colonized CF patients
In a large clinical study investigating the effect of sinus surgery on disease progression, a number of CF patients from the Copenhagen CF Center underwent sinus surgery. The procedures were performed as a standard computer-assisted functional endoscopic sinus surgery, where the maxillary sinuses and the ethmoidal sinuses as a standard were targeted and opened widely. Multiple samples were collected for cultivation including nasal secretions, pus, mucosal tissue, polyps and bone. Colonization and genotype profile patterns of P. aeruginosa in the airways of 45 children enrolled in the Copenhagen CF Center in the period from the beginning of 2005 until conclusion of the study in July 2009 have been investigated. Results are listed for each patient containing P. aeruginosa-positive sputum samples within the study period labeled with sample date and genotype (Supplementary Table S1). Genotypes of P. aeruginosa were determined using the AT-Biochip (Clondiag) method based on SNPs (Wiehlmann et al., 2007). This identification method was further supported by performing pulsed field gel electrophoresis of most clinical isolates, and in all cases identical conclusions concerning clonal relationships were obtained (not shown). The genotypic analysis allowed us to investigate if (1) each patient was colonized by a unique genotype of P. aeruginosa, (2) if the patient carried more than one genotype and (3) if the same genotype colonized both upper and lower airways in the patient.
In concordance with previous reports (Burns et al., 2001; Munck et al., 2001; Jelsbak et al., 2007), the majority of the children had acquired unique genotypes suggesting initial lung colonization from environmental sources rather than transmission between patients in our CF Center, the latter being restricted owing to cohort isolation (Høiby and Pedersen, 1989). However, eight specific genotypes were isolated from more than one patient (indicating transmission or common sources of colonization), and 9 of 45 children had more than one genotype isolated from their sputum. On the basis of the colonization pattern and genotype profiles, the CF patients could be divided into three groups: (a) Patients with single or multiple events of short colonization periods (<6 months) followed by eradication. Each colonization event was with a unique genotype. This group includes patients with new colonization events within the last 6 months before conclusion of study. (b) Intermittently colonized patients with multiple recurrent colonization events (usually separated by several months) with the same genotype of P. aeruginosa and a low number of precipitating antibodies (<2) indicating the absence of chronic lung infection (Høiby, 1974, 1977). (c) Patients with a rapid development of chronic lung infection with increasing numbers of precipitating antibodies (⩾2). A total of 31 patients (69%) belonged to group A with successful eradication after aggressive antibiotic treatments, and only three patients (7%) progressed directly to chronic infection (group C). In all, 11 patients (24%) belonged to group B and it is a primary hypothesis here, as suggested by several reports (Munck et al., 2001, 2003; Jelsbak et al., 2007; Mainz et al., 2009) that the bacteria were hiding in the paranasal sinuses. At conclusion of the study, 6 of the 11 group B patients had developed chronic infection after a period of intermittent colonization (Supplementary Table S1).
Cultivation and identification of bacteria were performed on sample materials from the six group B patients (B11, B12, B22, B28, B34 and B42 (had surgery twice)). We additionally included a young CF patient (B13, had surgery twice) who had been chronically infected for approximately 6 years. Thus, a total of nine events of sinus surgery performed on seven patients are referred to in this study (Supplementary Table 2).
The P. aeruginosa genotype (determined as SNP genotype from AT-Biochip assays supported by pulsed field gel electrophoresis) was unique for each patient, and the same specific genotype could be found consistently in all P. aeruginosa-positive sputum samples in each patient as well as in the patient's sinus samples (Table 2). Patient B13 is the only exception as three genotypes were present in the sputum samples, of which only two were present in the sinus samples; however, the third genotype was only isolated once from sputum. For six of the seven patients (incl. B13) P. aeruginosa was identified in the paranasal sinus samples. In patient B12, only the left side of the nose was cultured and no P. aeruginosa was found in the cultured sinuses. In the other eight events, both left and right side was cultured, an average of four sinus cavities were cultured, each with several samples and P. aeruginosa was found in all cultured sinus samples.
Consequently, genotype identity exists between the lower airways and the sinuses supporting the hypothesis that the paranasal sinuses constitute a colonized niche in the CF airways. The results further suggest that the sinuses represent a protected environment in which bacteria may survive even after their eradication from the lungs (Low et al., 2001). Despite the genotypic identity of the isolates derived from each patient, it was noted, however, that upon plating of individual clones, a clear diversity was apparent among the obtained bacterial colonies. It was therefore decided to investigate further this apparent diversity of the bacterial populations.
The paranasal sinuses harbor diverse bacterial subpopulations
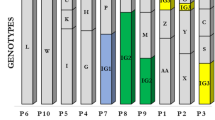
Colony morphology (morphotype) diversity of the sinus isolates was assessed for each patient by microscopy as described in Materials and methods. Each morphotype could be stably maintained after repeated growth, and the SNP genotype was determined for all variants. All available sputum samples from the lower airways were similarly examined for different morphotypes, and sinus and sputum populations were carefully compared to determine the number of identical morphotypes isolated until the point of surgery (Table 2). A strain list of all isolates (including morphotypes) is displayed in Supplementary Table S2, and the morphotype diversity found in two of the patients (B22 and B34) is shown in Figure 1.
Colony morphotypes of sinus and lower airway isolates from patients B22 and B34. Diversity in colony morphotypes is seen in and often shared between the paranasal sinuses and lower airways. (a) All morphotypes from patient B22. (*) A morphotype and genotype identical to B22-9_H was isolated again in June 2009 (B22-10_H). (b) All morphotypes from patient B34. A representative of each morphotype is displayed with sample date and isolate name.
Interestingly, substantial diversification of P. aeruginosa was observed in the sinus cavities. In two patients (B11 and B13), 10 or more morphotypes were found and in average 6.7±2.9 (mean±s.d.) morphotypes per patient were isolated from the sinuses. For the lower airway samples, the observed diversity was in average 5.3±1.2 (mean±s.d.) morphotypes per patient. These numbers most likely represent minimum estimates owing to the partial sample material and limitations in screening method. The overlap of morphotypes between the sinuses and the lower airways was high; on an average 3.2±1.2 (mean±s.d.) or 59% of the morphotypes found in the lower airways were also cultivated from the paranasal sinuses. Patients B22 and B34 both had three morphotypes shared between the sinuses and the lower airways (Figure 1). Many of the lower airway morphotypes that were not found in the sinuses belonged to the very early isolates from the sputum. In addition, the population in the left and right sinus cavities on each side of the nose in most patients had evolved into distinct morphotype populations (Table 2). In one patient (B28), a few morphotypes were found in both sides, but it is not known if this was a result of cross-contamination during surgery, as in this case the same instruments were used in the right and left sinuses. Patient B13 who was chronically infected at the time of the sinus surgery carried different genotypes in each side of the sinuses, a result that was confirmed at the second surgery 8 months later. The genotype found in the left sinuses was the only type recovered from the sputum samples until 2006, after which only the genotype from the right sinuses was identified in sputum (Supplementary Table S2). The apparent diversity of the P. aeruginosa populations in the sinus samples from the investigated group of patients strongly suggest that evolutionary changes occur, and that some of the evolved morphotypes may be similar to those documented from lung samples. These observations made us hypothesize that the bacteria colonizing the sinuses evolve to become distinct lineages in the CF upper airways, and that these lineages migrate to the lungs, where they may establish and become persistent. In the following sections, we will present data supporting this hypothesis.
Relationships between morphotype, phenotype and genotype
The finding that the sinuses and the lower airways are often colonized by identical P. aeruginosa morphotypes suggests that the two compartments harbor highly similar or identical lineages. If the isolates indeed are identical, it would be evidence of transfer between the sinuses and the lower airways. Therefore, phenotypic and genetic profiles of all isolates from patients B22 and B34 were compared. Specifically, phenotypes known to change during chronic infections in CF patients (Mahenthiralingam et al., 1994; Demko et al., 1995; Smith et al., 2006) such as motility, quorum sensing, secreted virulence factors and biofilm formation were analyzed, and the lasR and mucA genes were sequenced for certain isolates. In general, it was found that each morphotype had a distinct phenotypic profile displaying variable reduction or loss of the analyzed traits (Table 3). In patient B22, all isolates from the left sinuses had the same insertion mutation in the lasR regulator gene, and all were quorum sensing negative in contrast to the isolates from the right sinuses. In patient B34, the isolates from the right sinuses were mucoid owing to mutations in mucA causing alginate overproduction, whereas the left side isolates were non-mucoid and mucA+.
In patient B22, morphotypes E, F and G were found both in the sinuses and lower airways. The two lower airway isolates, B22-7_F and B22-8_G, were collected within the last 3 months before sinus surgery, and they displayed phenotypic profiles identical to the sinus isolates B22-sin_F and B22-sin_G, respectively (motility and biofilm formation values were within the standard deviation for each morphotype) (Table 3). The IS insertion causing a lasR mutation found in the entire left sinus population was also identified in the lower airway isolate B22-7_F, further suggesting a common origin. The E morphotypes had identical profiles as well, except for an 1.5-fold difference in biofilm formation capacity.
In patient B34, the shared morphotypes were B, M (mucoid) and SCV. All M isolates showed very similar phenotypic profiles with minor differences, which could in some part be due to instability of this phenotype, as it was observed to revert during experiments (DeVries and Ohman, 1994). However, the last mucoid morphotype isolated from the lower airways before surgery (B34-5_M) was not different from the mucoid sinus isolate B34-sin_M with respect to the tested phenotypes (within the standard deviation), and only six genes were differentially expressed at the transcriptomic level, P<0.05 and ⩾2-fold change (Supplementary Table S3). The sinus and lower airway mucoid isolates all had the same two mutations in the mucA gene (T261C, 262 C insert), except the first mucoid isolate, B34-2_M, which had a one base pair deletion in mucA (585 ΔA), suggesting that this lineage became outcompeted and possibly went extinct. Both the B and SCV morphotype isolates from the sinuses and lower airways (B34-4_B vs B34-sin_B and B34-6_SCV vs B34-sin_SCV) displayed identical phenotypic profiles (within the standard deviation). Thus, the morphotypes found in both the sinuses and lower airways of patients B22 and B34 were highly similar or identical at a phenotypic and genetic level documenting transfer between the two compartments. Intra-morphotype diversity seemed negligible as phenotypic profiling of several isolates of the same morphotypes from B22 showed that they were in general very similar, although small differences could occur (data not shown). The strong correlation between morphotypes and phenotypes shows that colony morphologies can be used successfully to screen population diversity, a method that has also been applied in several other studies (Rainey and Travisano, 1998; Boles et al., 2005; Hansen et al., 2007). The data also suggest that sinus and lower airway isolates from other patients having identical colony morphologies may reflect identical or at least closely related variants, suggesting transfer between the two compartments for these patients as well. Most importantly, however, the phenotypic and genotypic characterization of several isolates indicate that diversification of the sinus bacterial population leads to the development of lineages with the potential of establishing chronic infections also in the lower airways of the patients.
Sinus isolates with increased antibiotic resistance from patient B34
The presence of antibiotic-resistant strains in the sinuses could implicate the sinuses as reservoirs for colonization of the lower airways with pre-adapted strains. Therefore, the level and mechanism of antibiotic susceptibility was investigated for isolates from patient B34 by determining MICs and global gene expression analysis. In these isolates, susceptibility to ciprofloxacin decreased during the infection (Figure 2). Isolate B34-2_M, the subsequent mucoid (M) isolates and the SCV isolates were moderately less susceptible than the initial B34-1_A isolate, while B34-sin_MB type isolates were found to be resistant. Notably, the highest levels of resistance for any isolate type were always associated with the sinus isolate of the respective type. Isolates chosen for gene expression analysis included the first isolate B34-1_A, the last B morphotype before surgery (B34-4_B), the last mucoid isolate before surgery (B34-5_M) and the mucoid sinus isolate B34-sin_M (see Figure 1b for morphotypes and Supplementary Table S3 for gene expression changes). All isolates showed increased expression of the mexCD-oprJ operon, 1.9–2.6-fold for B34-4_B, 1.4–1.9-fold for B34-5_M and 5–10-fold for B34-sin_M relative to the first isolate B34-1_A (Supplementary Table S3). Overproduction of the MexCD-OprJ efflux pump is known to reduce susceptibility to antibiotics such as ciprofloxacin (Poole, 2004). Both MIC and gene expression data indicate that at least two or more separate events of increased ciprofloxacin resistance occurred in the B34 lineage. It is not certain in which part of the airways the increase in resistance evolved, yet interestingly the mucoid lower airway isolate B34-7_M shows no further increase in ciprofloxacin resistance in contrast to what was found for B34-sin_M isolated at an earlier time point. It is possible that the increased level of mexCD-oprJ expression observed in B34-sin_M evolved in the sinuses, and therefore that the increased resistance was selected for in the sinuses. It should also be noted from the transcriptomic data presented in Supplementary Table S3 that genes known to display reduced expression in isolates of P. aeruginosa from chronically infected CF patients (Yang et al., 2011), such as genes connected to motility, type III secretion and other virulence factors, also showed reduced expression in the B34 sinus isolates, supporting the hypothesis that the sinus populations evolve towards phenotypes associated with chronic infections.
Ciprofloxacin resistance profile of sinus and lower airways isolates of patient B34. The MIC of ciprofloxacin for patient B34 isolates was determined by E-test. Isolates are grouped into morphotype lineages with B34-1_A as a model ancestor. An increase in resistance was observed for the mucoid isolates, of which B34-sin_M has the highest level of resistance. The B morphotype and SCV found in the left side of the sinuses had increased resistance to the same level as the previously isolated identical morphotypes from lower airways. Patient isolates obtained after sinus surgery are not included. At least two replicate experiments were performed for each strain.
Parallel adaptive mutations in sinuses and lower airways
Screening of sinus isolates from all patients revealed the occurrence of mutations and phenotypes, which are frequently observed in CF lung isolates. An intriguing finding was mutations in mucA and lasR and the isolation of SCVs, as these genetic changes are known to promote disease progression in relation to chronic lung infection (Deretic et al., 1995; Haussler et al., 1999; Parad et al., 1999; Hoffman et al., 2009). To determine the frequency of these mutants in the sinuses, we screened the phenotypes of all sinus morphotypes isolated in the additional patients (Table 4). Colony morphology studies documented the presence of SCVs in the sinus populations, but CF lung SCV isolates have some additional characteristics besides the small colony form (Haussler et al., 1999; Starkey et al., 2009). Sinus SCV isolates were therefore further analyzed for these specific phenotypes: clumping in liquid culture, decreased motility, enhanced biofilm formation and antibiotic resistance (Figure 3). Susceptibility towards three antibiotics routinely given to the patients, ciprofloxacin, tobramycin and colistin, were tested. Two of the SCVs were less susceptible to tobramycin when compared with the initial isolate, whereas one strain in addition to (B28-sin_SCV) displayed decreased susceptibility to ciprofloxacin. No SCVs displayed increased susceptibility to colistin. As the potential SCVs also displayed clumping, decreased motility and enhanced biofilm formation, it could be concluded that four out of six patients possessed SCVs in the sinus population. Morphotypes that did not produce the Las-dependent signaling molecules were only observed in two patients (B22 and B34). In addition to patient B34, a mucoid variant was also present in the sinuses of patient B11, whereas loss or severe reduction in twitching and swimming motility was observed for some sinus isolates from all six patients (Table 3 and Supplementary Table S2). The finding of multiple phenotypes associated with chronic CF lung isolates among the sinus populations suggests that these phenotypes are adaptive both in the lower airways and in the sinus environment.
SCVs from the paranasal sinuses display similar characteristics as SCVs from chronic lung infection. (a) Colony morphology of first isolate and SCV from patients B34, B28, B42 and B11. SCVs were routinely grown on LB and not PIA amp plates owing to a higher instability on the latter. On PIA amp plates, the morphology of SCVs resembled isolate B42-sin_SCV. (b) Motility was generally reduced or lost in SCVs when compared with the first isolate. Swimming motility was also reduced in all SCVs; however, some activity still remained (except for B34-sin_SCV). Motility zone diameters are presented as mean±s.d. for at least three replicates. White bars, twitching motility; gray bars, swimming motility; black bars, swarming motility. (c) Biofilm formation abilities in microtiter plates were significantly increased for all SCVs. Data are presented as mean±s.d. for seven replicates. (d) The antibiotic resistance profiles of the SCVs were strain specific. Resistances to ciprofloxacin (black bars) and tobramycin (white bars) were determined by E-test. At least two replicate experiments were performed for each strain.
Transfer of P. aeruginosa between sinuses and lower airways
The results presented above strongly suggest that P. aeruginosa migrates between sinuses and lower airways, although without addressing the directionality. The fact that most patients have distinct morphotype populations in each side of the sinuses indicate little or no mixing of the two sinus populations. If the sinuses were occasionally seeded and invaded with different morphotypes that had evolved in undetectable populations residing in the lower airways (or oropharynx), one would expect to find mixed and more similar populations in both sides of the sinuses. Sinus surgery also seems to affect the lower airway population as one patient (B22) had no P. aeruginosa-positive sputum samples for almost 1 year and 6 months after sinus surgery, and the following colonization occurred with a new (different) genotype, suggesting a complete eradication of the previous genotype. Patient B42 also did not have recurrent colonizations after the second surgery until conclusion of the study in July 2009, and small reductions in lower airway colonization frequencies were seen for a couple of the other patients (Supplementary Table S1). These findings suggest that the sinus populations are intermittently seeding the lower airways, and that removal of these may offer a temporary relief of sinus symptoms and possibly lung colonization events (HK Johansen, personal communication; Jones et al., 1993; Nishioka et al., 1995).
On the basis of colony morphology and other phenotypic characteristics, we have constructed a model for a tentative development of the sinus populations of P. aeruginosa in patient B22 (Figure 4). According to the phenotypic data, it seems that two different phenotypic lineages were found among the longitudinal lower airway samples, each lineage correlating with a specific subpopulation from the right or left side of the sinuses. The most likely scenario for the evolutionary trajectory of the B22 isolates (Figure 4) shows that in one of the apparent lineages several phenotypes were lost or reduced in a step-wise manner over time, whereas the other lineage is almost unchanged for the tested phenotypes. Probably the two lineages branched even before the first isolate as none of the early evolutionary events are shared. Indeed, the point of divergence of the lineages could be the time of segregation into left and right sinus cavities and hence the divergent phenotypes between each side could reflect evolutionary events actually occurring in the sinuses. The lower airway isolates could consequently represent ‘snapshots’ of the sinus populations at a given time in accordance with the possibility that migration of P. aeruginosa in the early colonization stages mainly occurs in a downward direction from the sinuses towards the lungs.
Reconstruction model of the evolution of sinus populations in patient B22. On the basis of the phenotypic and genetic profile, relatedness of isolated variants from patient B22 could be estimated. All isolates are of identical SNP genotype. The profiles of lung isolates reveal that they likely represent previous stages in the evolution of variants from the sinus populations. The observed evolutionary events of a very common left sinus morphotypes (B22-sin_F) could, for example, be followed in a step-like manner through the longitudinal lower airway isolates. This was also the case in the right sinus population; however, only one major change was observed. Swarming motility was already lost in the first lower airway isolate (B22-1_A), but not in all successive lower airway isolates, suggesting that the first isolate from this patient does not fully represent the first colonizing strain (termed ‘WT’) and that segregation into each side of the sinuses happened before the collection of first lower airway isolate. As the WT phenotype is unknown, first isolate B22-1_A was chosen as a reference strain for phenotypic comparison, except for swarming motility where B22-6_E was used as reference strain. Each vertical step represents an evolutionary event that occurred at some point before the sample was collected (see timeline) and all major phenotypic changes observed (relative to first isolate) at that point are shown. ↓, considerably reduced (>70%) or lost phenotype.
Discussion
The initial hypothesis of this study was that the paranasal sinuses of CF patients constitute an alternative colonization site with reduced chances of antibiotic- and immune response-mediated clearance. This was supported by the findings that most CF children carry P. aeruginosa populations in their sinuses, and in all cases studied here the genotypes of these were the same as those colonizing the lungs. Surprisingly, it turned out that the sinus bacterial populations diversify into mixed populations of clonally related variants that migrate to the lungs and establish colonization with pre-adapted lineages, which eventually may result in chronic infection. It is practically and ethically impossible to prove a definite causal relationship between sinus colonization and chronic lung infection, and that the infection originates from the patients' sinuses. However, our study provides arguments, which support the proposition that the sinuses constitute a focus site for early P. aeruginosa adaptive evolution directing the colonization towards chronic lung infection.
Some intermittent colonization in CF patients end after aggressive antibiotic therapy as P. aeruginosa cannot be cultured from the sputum upon subsequent microbiological examination. The interpretation of this is that the bacteria are eradicated from the combined airways of the treated patients, and any later colonization events thus derive from new environmental strains. Depending on the success of the treatment regimen, this situation can continue for many years, suggesting that the P. aeruginosa population size and time of residence in the airways do not allow adaptive mutations important for the establishment of chronic infection to evolve. Other intermittent colonization events, however, display a different pattern with multiple recurrent colonization events (usually separated by several months) with the same genotype. In these patients, sinus infections with P. aeruginosa may provide a source for the occasional detection and low number of P. aeruginosa in the sputum samples. Our results show that the second-site colonization events in the sinuses, often being more persistent than those in the lungs, may provide extended opportunities for evolution of the bacteria towards phenotypes, with greatly increased potential of creating chronic lung infections as well. Although it has long been known that the microbiotas of the upper and lower airways are similar in chronically infected CF patients, only a small number of studies have investigated the possibility of cross-infections between the paranasal sinuses and the lungs (Dosanjh et al., 2000; Muhlebach et al., 2006; Mainz et al., 2009). In a large cohort study, almost half of the patients with a history of chronic P. aeruginosa lung infection also had an infection with the same genotype in the upper airways (Mainz et al., 2009). Identical genotypes in the sinus and bronchoalveolar lavage (lung) samples have also been recovered from one study of CF children with chronic sinusitis, and the frequency of identical genotypes increased from 9% of children up to 8 years old to 30% of children more than 8 years old (Muhlebach et al., 2006).
In our study, the sinuses were found to display substantial diversity in the P. aeruginosa population, and in most cases the population had segregated into two distinct populations on each side of the nose. Although we cannot totally exclude that specific subpopulations of bacteria from the lungs are transmitted to individual sides of the sinuses from time to time, it seems unlikely that a specific population in the lower airways would always be transferred to a specific sinus side, in particular during the period of intermittent lung colonization, during which antibiotic treatment effectively removes the lung populations of P. aeruginosa. Therefore, these results suggest that the direction of migration is mainly downwards at the early stages of infection. If it is assumed that recurrent intermittent lung colonization with the same genotype is associated with persistent sinus colonization, ours and other studies show that around 25% of the CF children may begin with a sinus colonization (Munck et al., 2001; Gibson et al., 2003; Doring et al., 2006), and more than half of all patients will eventually be affected (Mainz et al., 2009).The study further shows that phenotypic and genotypic changes observed in sinus isolates of P. aeruginosa are similar or identical to those often reported for isolates from chronic lung infections in CF patients. Several of the mutations and phenotypes that we have observed among sinus isolates, evolved in parallel in the different patients, including those conferring loss of motility and quorum sensing signals (virulence), alginate overproduction, antibiotic resistance and increased biofilm formation (SCVs). (Cabral et al., 1987; Mahenthiralingam et al., 1994; Haussler et al., 1999; Smith et al., 2006; D'Argenio et al., 2007; Starkey et al., 2009; Hoffman et al., 2010). Most of them are generally thought to be beneficial also in the lung environment through the development of resistance to host defenses and antibiotics or to optimized utilization of available nutrients. The effective removal of lung-associated P. aeruginosa during intermittent colonization of young CF patients and the consequential directional dominance of migration from sinuses to lungs strongly support our proposition that the observed diversity of the sinus-associated bacterial populations is predominantly caused by localized adaptive evolution in the sinuses.
The suggested role of the paranasal sinuses as persistent reservoirs for evolving bacterial populations may be associated with differences in the sinus and lung environments. Although the physiological properties and environment of the paranasal sinuses and conductive bronchi (where the lung infection is mainly residing) are likely to be comparable, as they have similar mucous lining and the same defect in CF transmembrane regulator, some differences in the immune response seem to exist. Lung infection with P. aeruginosa usually elicits stimulation of the innate immune system (inflammation) and an increase in precipitating antibodies against P. aeruginosa (Høiby, 1977), but the CF children from this study had colonization of the sinuses for long periods without an elevated systemic (immunoglobulin G) antibody response. Instead, it was recently found that immunoglobulin antibodies were prevalent and inflammation low, as also indicated by low numbers of neutrophils associated with the bacteria in the sinus samples (HK Johansen, personal communication).
Another feature of the sinuses is the detention of thickened mucus due to mucosal edema and closing of the sinus ostia (connection to the nose). It is very likely that the nutritional conditions in a partially or fully concealed sinus cavity are different from the lung environment where the sputum is produced and replaced continuously. Airflow into the sinus cavities can be reduced and lower oxygen tension in the sinuses and anoxic conditions on the sinus mucosa are seen in CF patients. These findings can vary from one side to the other of the noses (Carenfelt and Lundberg, 1977; Aanaes et al., 2011). Hence, bacteria may be locally faced with nutrient limitations and possibly starvation, in particular in some of the smaller cavities such as the ethmoids.
Nutrient limitation including low oxygen tension may be part of the conditions that facilitated the generation of mucoid variants (Terry et al., 1991, 1992) in two of six sinus-colonized patients, possibly in combination with other advantages such as biofilm formation and protection from phagocytosis facilitated by the alginate exopolysaccharide (Cabral et al., 1987) and evasion of host defense via downregulation of motility and virulence factors (Rau et al., 2010). Nutrient availability may also explain, in combination with antibiotic treatment, the selection of the lasR mutant population found in the left side of the sinuses of patient B22. Recent findings have shown that lasR mutants exhibit a dramatic metabolic shift with decreased oxygen and increased nitrate utilization and in addition they display increased resistance to antibiotics such as ciprofloxacin and tobramycin (Hoffman et al., 2010). Therefore, the lasR mutation is also likely to confer increased fitness in the sinuses.
Partially obstructed sinus cavities lead to reduced access of administered antibiotics, further resulting in lower antibiotic load compared with the lower airways. Sublethal antibiotic concentrations could provide opportunities both for survival as well as sufficient time for evolving increased antibiotic resistance. Altogether, the environment of the sinuses seems to differ from that of the lower airways in several important aspects: The immune response is apparently much weaker in the sinuses, the antibiotic bioavailability is low, antibiotics may be less effective owing to the physiological state of the bacteria in the sinuses and the environmental conditions in the sinuses may stimulate the occurrence of antibiotic-resistant mutants. In essence, what is important for the clearance of intermittently colonizing P. aeruginosa in the CF lungs is only partially functional in the sinuses, providing opportunities for the bacteria to adapt through evolution of resistance mechanisms with severe impacts on subsequent treatment possibilities in relation to bacteria migrating from the sinuses to the lungs.
Our study suggests that the paranasal sinuses can be an evolutionary ‘nest’ in early colonizations, where the bacteria are diversifying, evolving antibiotic resistance and other phenotypes associated with adaptation to the CF airways in general. From there the population is intermittently colonizing the lungs and may ultimately cause a chronic lung infection. On the basis of our findings, we suggest that the paranasal sinuses could have an important role for some CF patients in the development of chronic lung infection, and that chronicity in such cases is in fact established before it is usually diagnosed in the clinic. A precise localization and diagnosis of the CF airway colonizations and infections are therefore crucial to target treatments. Early diagnosis and successful treatment of P. aeruginosa colonizations in the paranasal sinuses could be an important therapeutic approach to prevent or delay transition to chronic lung infection and ultimately prolong the life of the patients.
References
Aanaes K, Rickelt LF, Johansen HK, von Buchwald C, Pressler T, Høiby N et al. (2011). Decreased mucosal oxygen tension in the maxillary sinuses in patients with cystic fibrosis. J Cyst Fibr 10: 114–120.
Andersen JB, Heydorn A, Hentzer M, Eberl L, Geisenberger O, Christensen BB et al. (2001). gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol 67: 575–585.
Boles BR, Thoendel M, Singh PK . (2005). Genetic variation in biofilms and the insurance effects of diversity. Microbiology 151: 2816–2818.
Brown MR, Foster JH . (1970). A simple diagnostic milk medium for Pseudomonas aeruginosa. J Clin Pathol 23: 172–177.
Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M et al. (2001). Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183: 444–452.
Cabral DA, Loh BA, Speert DP . (1987). Mucoid Pseudomonas aeruginosa resists nonopsonic phagocytosis by human neutrophils and macrophages. Pediatr Res 22: 429–431.
Carenfelt C, Lundberg C . (1977). Purulent and non-purulent maxillary sinus secretions with respect to pO2, pCO2 and pH. Acta Otolaryngol 84: 138–144.
Coste A, Gilain L, Roger G, Sebbagh G, Lenoir G, Manach Y et al. (1995). Endoscopic and CT-scan evaluation of rhinosinusitis in cystic fibrosis. Rhinology 33: 152–156.
D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE et al. (2007). Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64: 512–533.
Dacheux D, Attree I, Toussaint B . (2001). Expression of ExsA in trans confers type III secretion system-dependent cytotoxicity on noncytotoxic Pseudomonas aeruginosa cystic fibrosis isolates. Infect Immun 69: 538–542.
Demko CA, Byard PJ, Davis PB . (1995). Gender differences in cystic fibrosis: Pseudomonas aeruginosa infection. J Clin Epidemiol 48: 1041–1049.
Deretic V, Schurr MJ, Yu H . (1995). Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol 3: 351–356.
DeVries CA, Ohman DE . (1994). Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176: 6677–6687.
Doring G, Taccetti G, Campana S, Festini F, Mascherini M . (2006). Eradication of Pseudomonas aeruginosa in cystic fibrosis patients. Eur Respir J 27: 653.
Dosanjh A, Lakhani S, Elashoff D, Chin C, Hsu V, Hilman B . (2000). A comparison of microbiologic flora of the sinuses and airway among cystic fibrosis patients with maxillary antrostomies. Pediatr Transplant 4: 182–185.
Gibson RL, Emerson J, McNamara S, Burns JL, Rosenfeld M, Yunker A et al. (2003). Significant microbiological effect of inhaled tobramycin in young children with cystic fibrosis. Am J Respir Crit Care Med 167: 841–849.
Gysin C, Alothman GA, Papsin BC . (2000). Sinonasal disease in cystic fibrosis: clinical characteristics, diagnosis, and management. Pediatr Pulmonol 30: 481–489.
Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB . (1983). Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun 42: 170–177.
Hansen SK, Rainey PB, Haagensen JA, Molin S . (2007). Evolution of species interactions in a biofilm community. Nature 445: 533–536.
Haussler S . (2004). Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol 6: 546–551.
Haussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I . (1999). Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis 29: 621–625.
Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR et al. (2002). Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148: 87–102.
Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW et al. (2009). Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8: 66–70.
Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M et al. (2010). Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathogen 6: e1000712.
Høiby N . (1974). Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol Microbiol Scand B 82: 541–550.
Høiby N . (1977). Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. A survey. Scand J Respir Dis 58: 65–79.
Høiby N, Frederiksen B . (2000). Microbiology of cystic fibrosis. In: Hodson ME, Geddes DM (eds). Cystic Fibrosis, 2nd edn. Arnold: London, United Kingdom, pp 83–107.
Høiby N, Frederiksen B, Pressler T . (2005). Eradication of early Pseudomonas aeruginosa infection. J Cyst Fibros 4(Suppl 2): 49–54.
Høiby N, Pedersen SS . (1989). Estimated risk of cross-infection with Pseudomonas aeruginosa in Danish cystic fibrosis patients. Acta Paediatr Scand 78: 395–404.
Holloway BW, Morgan AF . (1986). Genome organization in Pseudomonas. Annu Rev Microbiol 40: 79–105.
Huxley EJ, Viroslav J, Gray WR, Pierce AK . (1978). Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med 64: 564–568.
Jelsbak L, Johansen HK, Frost AL, Thogersen R, Thomsen LE, Ciofu O et al. (2007). Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun 75: 2214–2224.
Jones JW, Parsons DS, Cuyler JP . (1993). The results of functional endoscopic sinus (FES) surgery on the symptoms of patients with cystic fibrosis. Int J Pediatr Otorhinolaryngol 28: 25–32.
Koch C . (2002). Early infection and progression of cystic fibrosis lung disease. Pediatr Pulmonol 34: 232–236.
Lam J, Chan R, Lam K, Costerton JW . (1980). Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun 28: 546–556.
Low AS, MacKenzie FM, Gould IM, Booth IR . (2001). Protected environments allow parallel evolution of a bacterial pathogen in a patient subjected to long-term antibiotic therapy. Mol Microbiol 42: 619–630.
Mahenthiralingam E, Campbell ME, Speert DP . (1994). Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun 62: 596–605.
Mainz JG, Naehrlich L, Schien M, Kading M, Schiller I, Mayr S et al. (2009). Concordant genotype of upper and lower airways P aeruginosa and S. aureus isolates in cystic fibrosis. Thorax 64: 535–540.
Muhlebach MS, Miller MB, Moore C, Wedd JP, Drake AF, Leigh MW . (2006). Are lower airway or throat cultures predictive of sinus bacteriology in cystic fibrosis? Pediatr Pulmonol 41: 445–451.
Munck A, Bonacorsi S, Mariani-Kurkdjian P, Lebourgeois M, Gerardin M, Brahimi N et al. (2001). Genotypic characterization of Pseudomonas aeruginosa strains recovered from patients with cystic fibrosis after initial and subsequent colonization. Pediatr Pulmonol 32: 288–292.
Nguyen D, Singh PK . (2006). Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci USA 103: 8305–8306.
Nishioka GJ, Barbero GJ, Konig P, Parsons DS, Cook PR, Davis WE . (1995). Symptom outcome after functional endoscopic sinus surgery in patients with cystic fibrosis: a prospective study. Otolaryngol Head Neck Surg 113: 440–445.
Parad RB, Gerard CJ, Zurakowski D, Nichols DP, Pier GB . (1999). Pulmonary outcome in cystic fibrosis is influenced primarily by mucoid Pseudomonas aeruginosa infection and immune status and only modestly by genotype. Infect Immun 67: 4744–4750.
Poole K . (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin Microbiol Infect 10: 12–26.
Pratt LA, Kolter R . (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30: 285–293.
Rainey PB, Travisano M . (1998). Adaptive radiation in a heterogeneous environment. Nature 394: 69–72.
Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF et al. (2010). Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12: 1643–1658.
Robertson JM, Friedman EM, Rubin BK . (2008). Nasal and sinus disease in cystic fibrosis. Paediatr Respir Rev 9: 213–219.
Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 103: 8487–8492.
Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A et al. (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191: 3492–3503.
Taccetti G, Campana S, Festini F, Mascherini M, Doring G . (2005). Early eradication therapy against Pseudomonas aeruginosa in cystic fibrosis patients. Eur Respir J 26: 458–461.
Taylor RF, Morgan DW, Nicholson PS, Mackay IS, Hodson ME, Pitt TL . (1992). Extrapulmonary sites of Pseudomonas aeruginosa in adults with cystic fibrosis. Thorax 47: 426–428.
Terry JM, Pina SE, Mattingly SJ . (1991). Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect Immun 59: 471–477.
Terry JM, Pina SE, Mattingly SJ . (1992). Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect Immun 60: 1329–1335.
Thomassen MJ, Demko CA, Boxerbaum B, Stern RC, Kuchenbrod PJ . (1979). Multiple of isolates of Pseudomonas aeruginosa with differing antimicrobial susceptibility patterns from patients with cystic fibrosis. J Infect Dis 140: 873–880.
Wahba AH, Darrell JH . (1965). The identification of atypical strains of Pseudomonas aeruginosa. J Gen Microbiol 38: 329–342.
Wiehlmann L, Wagner G, Cramer N, Siebert B, Gudowius P, Morales G et al. (2007). Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 104: 8101–8106.
Yang L, Haagensen JA, Jelsbak L, Johansen HK, Sternberg C, Høiby N et al. (2008). In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J Bacteriol 190: 2767–2776.
Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH et al. (2011). Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci USA 108: 7481–7466.
Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M et al. (2009). Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother 53: 2432–2443.
Acknowledgements
We gratefully acknowledge Matthew Parsek and Liang Yang for experimental assistance and we also thank the nurses and doctors at the Copenhagen CF Center for their help in recruiting and preparing the patients for surgery. This work was supported by grants from the Danish Research Agency and the Lundbeck Foundation to SM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Hansen, S., Rau, M., Johansen, H. et al. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J 6, 31–45 (2012). https://doi.org/10.1038/ismej.2011.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.83
Keywords
This article is cited by
-
Bacteriological Profile of Chronic Rhinosinusitis and Adenotonsillitis: Evaluating the Role of Biofilm Production and Multidrug Resistance
Indian Journal of Otolaryngology and Head & Neck Surgery (2023)
-
The difference in pathogenic bacteria between chronic rhinosinusitis in patients with and without Sjogren’s syndrome: a retrospective case–control study
BMC Infectious Diseases (2022)
-
Evolutionary highways to persistent bacterial infection
Nature Communications (2019)
-
Genetically diverse Pseudomonas aeruginosa populations display similar transcriptomic profiles in a cystic fibrosis explanted lung
Nature Communications (2019)
-
Evolutionary trade-offs associated with loss of PmrB function in host-adapted Pseudomonas aeruginosa
Nature Communications (2018)