Abstract
It has been long debated as to whether marine microorganisms have a ubiquitous distribution or patterns of biogeography, but recently a consensus for the existence of microbial biogeography is emerging. However, the factors controlling the distribution of marine bacteria remain poorly understood. In this study, we combine pyrosequencing and traditional Sanger sequencing of the 16S rRNA gene to describe in detail bacterial communities from the deep Arctic Ocean. We targeted three separate water masses, from three oceanic basins and show that bacteria in the Arctic Ocean have a biogeography. The biogeographical distribution of bacteria was explained by the hydrography of the Arctic Ocean and subsequent circulation of its water masses. Overall, this first taxonomic description of deep Arctic bacteria communities revealed an abundant presence of SAR11 (Alphaproteobacteria), SAR406, SAR202 (Chloroflexi) and SAR324 (Deltaproteobacteria) clusters. Within each cluster, the abundance of specific phylotypes significantly varied among water masses. Water masses probably act as physical barriers limiting the dispersal and controlling the diversity of bacteria in the ocean. Consequently, marine microbial biogeography involves more than geographical distances, as it is also dynamically associated with oceanic processes. Our ocean scale study suggests that it is essential to consider the coupling between microbial and physical oceanography to fully understand the diversity and function of marine microbes.
Similar content being viewed by others
Introduction
Microbes dominate the ocean in terms of abundance, diversity and metabolic activity (Azam and Malfatti 2007). Marine bacteria mediate fluxes of matter and energy and have a critical role in driving the major biogeochemical cycles (Karl, 2002). Although microbes have an essential role in ecosystem functioning, very little is known about the factors structuring marine community distribution. Microbial communities have been described as stratified with depth (Giovannoni et al., 1996; Field et al., 1997; Karner et al., 2001), and depth has until recently been considered as the main factor explaining differences in marine microbial community composition (DeLong et al., 2006; Pham et al., 2008). Light availability (irradiance) is thought to be the main abiotic factor structuring communities in the euphotic zone (Giovannoni and Stingl, 2005). Dark ocean communities, however, are not homogenous (Hewson et al., 2006; Teira et al., 2006), suggesting that other key factors besides irradiance influence vertical microbial community structure. Latitude has recently been proposed as an important factor determining surface microbial diversity (Pommier et al., 2007; Fuhrman et al., 2008), but other factors may control bacterial communities in the deep dark ocean.
The dark ocean comprises the water below 200 m, including the mesopelagic (200–1000 m depth) and bathypelagic (1000–4000 m depth) zones and represents the largest biome on earth (70% of the global ocean's volume). Sun irradiance does not reach deeper waters but, nevertheless, accumulating data suggest that they harbor diverse and active microbial communities (Fuhrman and Davis, 1997; Karner et al., 2001; Teira et al., 2006; Hansman et al., 2009). These communities contain potentially novel phylogenetic diversity and metabolisms, but their role in the oceans remains poorly understood. Deep bacterial communities remain less studied than surface communities, and to our knowledge, in the Arctic Ocean, the composition of deep bacteria communities has never been described.
The oceans are not uniform, but made up of regionally formed water masses, with distinct temperature and salinity characteristics, which move around the globe over different spatial scales (Stommel, 1958; Broecker, 1991). This thermohaline circulation has global significance for life on earth (Broecker, 1997), and communities of large plankton such as cnidarians are known to be structured by water masses (Hosia et al., 2008). Microbes are also influenced by the hydrography of the ocean, but that aspect of microbial oceanography remains poorly studied. Nevertheless, recent evidence suggests a link between microbial community composition and water masses (Agogue et al., 2008; Hamilton et al., 2008; Varela et al., 2008b; Galand et al., 2009b). Water masses could be a key factor explaining microbial biogeography, but this hypothesis remains to be tested at an ocean scale.
In this study, we hypothesized that deep marine bacterial communities have a biogeography that can be associated with water masses, that is, different water masses harbor different bacterial communities. We tested this hypothesis by targeting three different geographical regions of the Arctic Ocean, including three major Arctic oceanic basins: the Canada Basin, the Eurasian Basin and Baffin Bay. We designed our study to encompass three well-defined types of deep Arctic waters from the twilight zone: the halocline, the Atlantic layer and the Baffin Bay intermediate water. We then targeted the hypervariable V6 region of the bacterial 16S rRNA gene by pyrosequencing, resulting in an exhaustive description of the communities. In addition, we used traditional Sanger sequencing of clone libraries to produce longer 16S rRNA gene sequences to obtain more precise phylogenetic affiliations for the majority of pyrosequencing reads.
Materials and methods
Sampling sites and procedure
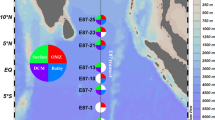
A total of 13 samples were collected during summer 2007 (from 12 July to 21 September 2007) from three major oceanic basins of the Arctic Ocean: the Canada Basin, the Eurasian Basin and the Baffin Bay Basin (Figure 1). Samples were collected to target three different deep arctic water masses: the halocline, the Atlantic layer and the Baffin Bay intermediate water (Table 1). Water masses were identified on the basis of their salinity–temperature characteristics (McLaughlin et al., 2004; Rudels et al., 2004; Tang et al., 2004). Samples from the Baffin Bay and Canada Basin (Figure 1) were collected from the Canadian icebreaker CCGS Louis St Laurent as part of the International Polar Year study ‘Canada's 3 Oceans and faunas project.’ Samples from the Eurasian Basin were collected from the R/V Victor Buynitskiy as part of the Nansen and Amundsen Basins Observation Systems. Water was sampled, microbial cells concentrated and DNA extracted as described earlier (Galand et al., 2009a).
Map of the Arctic Ocean showing the position of the sampling sites in the Canada Basin, Eurasian Basin and Baffin Bay, and the typical circulation of intermediate depth waters to 1700 m (adapted from Jones et al., 1995). Details on stations and sample depths are given in Table 1.
DNA extraction and pyrosequencing
The bacterial hypervariable V6 region of the 16S rRNA gene was amplified using primers specific for bacteria containing a pool of five forward and four reverse primer sets targeting the region between nucleotide 967 and 1046 (Escherichia coli numbering) (Huber et al., 2007). The final amplicons were sequenced at the Josephine Bay Paul Center of the Marine Biological Laboratory (Woods Hole, MA, USA) with a 454 Life Sciences GS20 sequencer producing 80–120 base long sequences or reads (Margulies et al., 2005). For each read, the primer bases were trimmed from the beginning and the ends and low-quality sequences were removed (Huse et al., 2007). Sequences were flagged as low quality when (i) they were less than 50 nucleotides (ii) the start of the sequence did not have an exact match to a primer sequence, (iii) the sequence contained one or more ambiguous nucleotides (Ns) or (iv) if the first five nucleotides of a tag did not correspond to the expected five nucleotide run key (used to sort the pyrosequencing reads). Pyrotag sequences have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession numbers SRX011229 to SRX011241.
Construction of clone libraries and phylogenetic analysis of cloned sequences
Nearly complete bacterial 16S rRNA genes were amplified from all samples with the bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) as previously described (Galand et al., 2008). PCR products were analyzed by gel electrophoresis, purified using the QIAquick PCR Purification Kit (Qiagen, Mississauga, Ontario, Canada) and cloned with TA cloning kit (Invitrogen, Burlington, Ontario, Canada). In total, we constructed 13 bacterial 16S rRNA gene libraries. For each library, positive clones were picked and inoculated into lysogeny broth (LB) media in 96-well plates. Bacterial clones were randomly chosen from each library and sequenced from both ends using the vectors universal primers. Suspected chimeras were checked using Bellerophon (Huber et al., 2004) and by using BLAST with sequence segments separately. The 16S rRNA sequences data obtained in this study have been archived in the GenBank database under accession numbers GQ337081–GQ337327.
The taxonomy of the nearly full-length sequences was assigned by the Sequence Match tool of the Ribosomal Database Project (RDP) II and by BLAST comparison with sequences archived in GenBank. Approximately 1400 bp sequences were aligned using MUSCLE (Edgar, 2004) and manually checked. Phylogenetic analyses were completed with the program PHYLIP (Felsenstein, 2004). DNADIST was used to calculate the genetic distances with Kimura-2 model, and the distance tree was estimated using FITCH.
Taxonomic identification of pyrosequencing reads and definition of operational taxonomic units
The taxonomic identification of the pyrosequencing reads (‘tags’) followed the approach proposed by Sogin et al. (2006). The tags were compared by BLASTN with a reference database of hypervariable region tags (RefHVR_V6, http://vamps.mbl.edu/) based on the SILVA database (version 95, Pruesse et al., 2007), and the 100 best matches were aligned to the tag sequences using MUSCLE (Edgar, 2004). A reference sequence or sequences were defined as those having the minimum global distance (number of insertions, deletions and mismatches divided by the length of the tag) to the tag sequence, and all reads showing the best match to the same reference V6 tag were grouped together as the same operational taxonomic unit (OTU) (‘best match’ definition, Dethlefsen et al., 2008; Galand et al., 2009a; Sogin et al., 2006). Taxonomy was assigned to each reference sequence with the RDP Classifier (Wang et al., 2007).
This pipeline was, however, not precise enough to classify all bacteria from the deep Arctic Ocean, for which a few sequence records are available in the databases. To increase the taxonomic resolution, pyrosequencing reads were compared against our nearly full-length sequences using BLASTN. This additional step improved the classification up to the class/order level for 30 760 sequences (9.8% of the sequences) that were originally classified at the domain level, and 55 550 sequences (17.7%) that were first defined at the phylum level.
Similarity between bacterial communities
To estimate community similarity among samples, we applied an hierarchical cluster analysis on the basis of the abundance of OTUs in the communities using Bray–Curtis similarity and a dendrogram inferred with the unweighted pair-group average (UPGMA) algorithm as implemented in the program PAST (v 1.90, Hammer et al., 2001). To determine the robustness of the clustering, data were subjected to bootstrapping with 1000 re-sampling.
Analysis of similarity (ANOSIM) statistics were used to verify the significance of the dendrogram clustering by testing the hypothesis that bacterial communities from the same cluster were more similar to each other than to communities in different clusters. A Bray–Curtis similarity matrix computed from the abundance of OTUs was used to generate one-way ANOSIM statistics with 10 000 permutations. Analyses were conducted with the program PAST (v 1.90, Hammer et al., 2001).
Diversity estimations and statistical analysis
Pyrosequencing reads were aligned with MUSCLE and pairwise distances calculated using the program Quickdist (Sogin et al., 2006). These pairwise distances served as input to DOTUR (Schloss and Handelsman, 2005) for clustering sequences into OTUs, generating rarefaction curves, and calculating the species richness estimator ACE and Chao1. The 97%, 94% and 90% similarity level between sequences were used for calculation of diversity estimators.
Sampling effort (number of sequences obtained for each sample) was not equally distributed, and samples with the highest number of sequences were expected to show a comparatively higher diversity. To normalize sampling effort across samples, sequences were randomly re-sampled through rarefaction analysis with the program DOTUR (Schloss and Handelsman, 2005), and estimates of richness were obtained from 15 400 sequences randomly drawn from each sample (Supplementary Table 6). A total of 15 400 sequences corresponded to the smallest sampling effort in our datasets (sample DAO_0009). Differences in richness between water masses were tested with one-way ANOSIM on a data set comprising richness estimators OTU, Chao and ACE calculated at 3% and 6% (n=78).
Similarity percentage was used to determine which individual OTU contributed most to the dissimilarity between samples. Principal component analysis was conducted with abundance data of OTUs using a variance–covariance matrix as implemented in PAST.
Variations in the abundance of bacterial taxa among water masses were compared by one-way ANOSIM calculated using PAST, and differences in abundance were considered significant when P<0.05.
Results
Similarity between bacterial communities and hydrography
Compositional similarity among the 13 bacterial communities was assessed by comparing the relative abundance and distribution of 313 827 bacterial 16S rRNA gene fragments (‘tags’). The cluster analysis revealed that communities were separated according to their water mass of origin. Water masses are described in detail in Supplementary results, and physical and chemical characteristics are given in Table 1 and Supplementary Figure 1.
At a similarity level >60%, we distinguished three major groups (A, B and C, Figure 2). Group A included all samples from the Atlantic layer of the Canada Basin. Group B consisted of the samples from the Halocline waters of the Canada Basin and the sample from Baffin Bay intermediate water. Group C included all samples collected from water masses of the Eurasian Basin. Within Eurasian waters (Group C), the two samples from the Atlantic layer grouped together separate from the halocline sample (Figure 2). Bootstrap values reached 100% for the three major groups and were otherwise always >50%, indicating a robust analysis. Two clusters remained at a similarity level >50%, one containing all samples from the Western Arctic water masses (Group A and B) and the other containing samples from the Eastern Arctic waters (Group C, Figure 2).
(a) Dendrogram representing the similarity between 13 bacterial communities from the Arctic Ocean. Clustering is based on a distance matrix computed with Bray–Curtis similarity from sequence abundance data. The dendrogram was inferred with the unweighted pair-group average algorithm (UPGMA). A, B and C indicate the three major groups identified for a similarity level >60%. Bootstrap values (in %) for 1000 replicates are given at the nodes. (b) Temperature–salinity diagram indicating water masses present in the Arctic Ocean. Circles mark the position of the samples and colors correspond to the three groups identified in the cluster analysis.
We used two different sequence similarity cut-offs for defining OTUs when comparing samples by cluster analysis. OTUs were first defined by grouping all sequences having the best match to the same reference sequence in the RefHVR_v6 database (‘best match’ definition, see Material and methods section for details), which roughly corresponded to a 94% identity between sequences in our study. The second definition was much more stringent, as each single sequence was considered as unique OTUs (100% identity cut-off). Cluster analyses run separately with the two OTU definitions yielded the same groupings, suggesting that the results were not biased by the stringency of OTU definition (data not shown).
The main purpose of our clone libraries was to verify taxonomic affiliations rather than to compare communities. Nevertheless, we also used the clone data set to run cluster analysis using the abundance of the nearly full-length 16S rRNA gene sequences. The resulting dendrogram had low bootstrap values at its nodes (Supplementary Figure 2), indicating the need for high numbers of clones to be analyzed for a reliable clone-based biogeographical study. Despite the low bootstrap values, the water mass-specific trends were comparable with pyrosequencing results (Supplementary Figure 2).
Diversity of deep Arctic communities
Out of the 313 827 bacterial tag sequences from pyrosequencing, there were 25 718 unique sequences that grouped into 4603 OTUs (‘best match’ definition) with a predicted total diversity of 7793 OTUs (‘best match’ definition, Chao1 estimator). Rarefaction analysis showed that the curve did not level off at the 97% cut-off, even for the samples with the greatest number of sequences (Figure 3), neither did the curves level off for the less stringent ‘best match’ OTU definition (Supplementary Figure S3), indicating a high bacterial diversity in the deep Arctic Ocean. Richness varied between samples with Chao1 values at the 97% threshold ranging from 1843 to 3530 (Table 2). The diversity of the Eurasian water masses (Group A, Figure 2) was significantly higher than the diversity of both the halocline and Atlantic layer of the Canada Basin (ANOSIM, P<0.02), but there was no differences in diversity between the halocline and Atlantic layer of the Western Canadian Arctic (Supplementary Table 6).
Overall, the most abundant groups in the deep Arctic Ocean were affiliated to the class Alphaproteobacteria (Figure 4), which represented 30% of the pyrosequencing tags. The second most abundant group was Deltaproteobacteria (19% of the tags), followed by Gammaproteobacteria (17%), the SAR406 cluster (9%) and Chloroflexi, mainly from the SAR202 cluster (5%). Other groups were rarer and many sequences were present at very low abundances with 17 543 sequences present only once.
Alphaproteobacteria were mostly represented by sequences belonging to the SAR11 cluster (67% of the tags were allocated to this phylogenetic class). The most abundant group within SAR11 (v6_AC000, 38% of Alphaproteobacteria sequences) were the SAR11-S1 cluster (Garcia-Martinez and Rodriguez-Valera, 2000), followed by the SAR11-D cluster (Supplementary Figure S4). Other Alphaproteobacteria also detected in the clone libraries were Rhodospirillales, Rhodobacterales and Rickettsiales. In addition, pyrosequencing identified sequences belonging to orders Caulobacterales, Kordiimonadales, Rhizobiales and Sphingomonadales.
Deltaproteobacteria belonged mostly to the SAR324 cluster (80% of the tags). One SAR324 OTU (V6_CY033, Supplementary Figure S5) was the single most abundant sequence overall. It alone represented 13% of all sequences from the entire data set, ranging from 10% to 17% in the different samples. Other Deltaproteobacteria OTUs were identified as Desulfobacterales and Myxococcales.
Gammaproteobacteria were more diverse. The most abundant group represented only 10% of the Gammaproteobacteria sequences and belonged to an uncultured cluster, previously named Arctic96BD-19 (Bano and Hollibaugh, 2002) or the GSO cluster (Lavik et al., 2009), containing a chemolithotrophic sulfide-oxidizer bacteria. The second most abundant group of Gammaproteobacteria sequences belonged to the ZD0417 cluster (Stevens and Ulloa, 2008) (Supplementary Figure S6). Other Gammaproteobacteria belonged to orders Oceanospirillales, Alteromonadales and Thiotrichales.
SAR406 sequences fell into five major clusters. The most abundant sequences belonged to the SPOTSMAY03_500m12 cluster (Supplementary Figure S7). Other abundant sequences were within the SAR406 cluster itself and SPOTSAPR01_5m105 cluster.
Chloroflexi sequences were mostly from the SAR202 cluster. The most frequent SAR202 sequences fell within the original SAR202/SAR307 cluster and under cluster 1 (Morris et al., 2004) (Supplementary Figure S8). We also detected Chloroflexi sequences belonging to classes Anaerolineae and Caldilineae.
Overall, the five most abundant groups of bacterial sequences had the same abundance across water masses (ANOSIM, P>0.05), but less abundant sequences were more variable in abundance (Figure 4). Actinobacteria and Acidobacteria sequences were less abundant in the Eurasian waters masses than in the Canada Basin halocline. Defferibacteres sequences were more abundant in the halocline of the Canada Basin. Planctomycetes, Gemmatimonadetes, Betaproteobacteria, especially from the OM43 cluster, and Lentisphaerae sequences were more abundant in the Eurasian waters (Figure 4).
Water mass-specific OTUs
We identified the OTUs (‘best match’ definition) contributing to the overall dissimilarity between water masses by using principal component analysis and similarity percentages statistics on the five most abundant classes/clusters of bacteria. The significance of the OTUs for explaining the differences was tested by ANOSIM.
Alphaproteobacteria. The 89 843 sequences identified as Alphaproteobacteria grouped into 960 OTUs. Among those OTUs, v6_AC000, v6_AC075, v6_AC028, v6_AC090 were the most important for explaining differences between water masses (Supplementary Table S1). The halocline contained significantly more v6_AC090 OTUs, the Atlantic layers had fewer v6_AC000 and more v6_AC028 OTUs, and both water masses from the Eurasian Basin had more v6_AC075 OTUs (Figure 5). In terms of taxonomy, the Eurasian deep waters had comparatively more SAR11 from the A21 clusters (according to nomenclature by Garcia-Martinez and Rodriguez-Valera, 2000) (Supplementary Figure S4), the Atlantic layer had more sequences from the SAR11 cluster IB (Suzuki et al., 2001) and the halocline had more sequences from SAR11-S1 and -D clusters (Garcia-Martinez and Rodriguez-Valera, 2000).
Principal component analysis (PCA) based on the relative abundance of Alphaproteobacteria operational taxonomic units (OTUs). The analysis comprises 89 843 sequences identified as members of Alphaproteobacteria. The most abundant OTUs (representing at least 1% of the sequences) are plotted at the top of the figure showing the PCA analysis. Grouping of samples corresponds to the assemblages determined by cluster analysis.
Deltaproteobacteria. The 58 592 sequences from Deltaproteobacteria grouped into 321 OTUs. The v6_CY033 was the most abundant OTU and represented more than 70% of all sequences. It was identified as belonging to the SAR324 cluster (Supplementary Figure S5) and had similar abundance in all water masses (ANOSIM, P>0.05). Deltaproteobacteria sequences did not separate according to the water masses, and the only significant difference in community composition was between the North American water masses and the Eurasian waters. The Eurasian waters were characterized by OTUs v6_CF143, whereas the North American waters had more v6_CF232 (Supplementary Table S2). All these OTUs were identified as members of the family Nitrospinaceae.
Gammaproteobacteria. The 47 625 Gammaproteobacteria sequences grouped into 825 OTUs. The Eurasian water masses were characterized by a greater proportion of v6_CY051 and v6_AD347 OTUs, the Atlantic layers had more v6_CG283, v6_CF951, and the halocline of the Canada Basin contained more v6_CG260 and v6_CY086 (Supplementary Table S3).
SAR406 and Chloroflexi. The 26 625 sequences belonged to the SAR406 cluster. SAR406 OTUs were distributed according to their water mass of origin. V6_CH277 OTU was present in larger quantities in the halocline waters of the Canada Basin, whereas OTUs V6_CH266 and V6_CO490 were more abundant in the Eurasian waters (Supplementary Table S4). The 14 077 sequences were assigned to the phylum Chloroflexi. OTUs v6_AG969 and v6_CV873 were more frequent in the halocline of the Canada Basin, v6_AZ408 and v6_AX625 were in large quantities in the Atlantic layer of the Canada Basin, and OTUs v6_CD831 and v6_CE057 were abundant in the Eurasian Basin water masses (Supplementary Table S5). All those Chloroflexi OTUs belonged to the SAR202 cluster (Supplementary Figure S8).
Discussion
The characterization of bacterial community structure across the deep Arctic Ocean revealed that communities had patterns of biogeography and that those patterns were related to water masses. Water masses thus appear as an important factor explaining the distribution of marine microbes in the deep layers of the ocean. Marine microbial communities are known to be vertically stratified (Giovannoni and Stingl, 2005) and depth has often been cited as the main factor explaining changes in community composition (Giovannoni et al., 1996; DeLong et al., 2006). Depth, as a proxy for irradiance penetration and barometric pressure, is certainly important for microbial zonation. Communities from the upper euphotic layers are mostly composed of phototrophs and heterotrophic organisms associated with those primary producers. The dark ocean, on the other hand, is inhabited by different communities consisting of mostly chemotrophs (Herndl et al., 2008; Hansman et al., 2009). In this study, we show that depth was not sufficient to explain microbial biogeography, especially within the twilight region of the dark ocean, and that water masses should be considered in efforts to understand the functional role and diversity of marine bacteria.
The oceanic circulation in the deep Arctic Ocean (see Figure 1) is well characterized (Jones et al., 1995) and is consistent with the main differences in community composition between the Canada Basin and the Eurasian Basin waters. The Eurasian Basin circulation follows a counterclockwise rotation bringing in new Atlantic water through the Barents Sea and Fram Strait. In the Canada Basin, a similar gyre system imports water from the upper Pacific Ocean that is fresher (less dense) and remains above old Atlantic deep water in the Canada Basin. The gyre system subsequently expels upper Arctic water through the shallow Canadian Archipelago. The deep waters circulating in the Eurasian and Canada Basins, separated by the Lomonosov Ridge, remain physically isolated from each other. The Lomonosov ridge rises up 3300–3700 m above the seabed and acts as a barrier separating the western and eastern sides of the Arctic Ocean. Only water from the upper water column can spill over from one basin to the other, and thus, the deep waters and associated bacteria of the Eurasian Basin remain separated from the deep Canada Basin waters, which are estimated to be ∼1000-years old (Macdonald and Carmack, 1991). Within each basin, bacterial communities also differed by depth-associated water masses. Halocline communities were always different from Atlantic layer communities, reflecting the different characteristics and origins of the water masses (Rudels et al., 2004). This was recently reported for Archaea, but the limited data set was less conclusive (Galand et al., 2009a). In this study, we found that similar communities were recovered from Baffin Bay waters and the Canada Basin halocline, consistent with water mass movements and circulation. In this situation, water from the Canada Basin enters Baffin Bay via Lancaster Sound after traveling through the Canadian Archipelago, and more directly through the Nares Strait between Greenland and the Canadian Ellesmere Island. That water has to pass over a shallow sill (230 m) and only halocline water reaches Baffin Bay and contributes to the formation of the Baffin Bay intermediate water (Rudels et al., 2004), which was the source of our DAO_ 0001 sample.
The change of bacterial communities with water mass suggests hydrographic control of bacterial biogeography. Two distinct explanations for the biogeographical distributions of larger plankton species have been proposed. The ‘barriers to dispersal’ models assume that the density characteristic of specific water masses constitute hydrographic boundaries that are obstacles for the dispersal of plankton (Spencer-Cervato and Thierstein, 1997). The ‘high dispersal’ models propose, in turn, that plankton disperse freely and continually but only maintain viable populations in favorable areas (Darling et al., 2004; Sexton and Norris, 2008). Both those models could be applied to explain the biogeography of bacteria in the deep Arctic Ocean. Water masses can remain isolated from each other because of physical barriers, such as density and oceanic ridges, isolating populations and allowing them to diverge through adaptation or random genetic drift. The ‘barrier to dispersal’ concept could explain the differences between the deep Canada Basin and Eurasian Basin bacterial communities. In contrast, even though we could not identify chemical parameters as structuring forces in our study, the observed biogeographical patterns could be due to the environmental heterogeneity represented by water masses with specific biotic and environmental properties. Different properties will result in different ecological niches and thus different bacterial communities. Those communities can be mixed when water masses blend. This process could account for the specific presence of the Betaproteobacteria OM43 cluster in the deep Eurasian waters. Deep water masses are not expected to contain the OM43 cluster, which is thought to be linked to phytoplankton blooms and productive coastal ecosystems (Giovannoni et al., 2008). Samples from the Eurasian Basin were, however, from a region where the halocline is formed by dense water that sinks toward the deeper layers of the ocean (Jones et al., 1995). Sinking water may draw down remnant surface bacteria episodically increasing the diversity of deep communities. Similarly, surface bacteria attached to sinking particles may be able to escape water mass barriers and reach greater depths. Once isolated in deep basins, genomic processes such as recombination and selection would operate over longer time scales (Konstantinidis and DeLong, 2008). The biogeography of marine bacteria is thus probably guided by complex interactions of communities isolated by water mass boundaries, communities migrating through frontal systems or sinking particles, and community selection through environmental filters and genomic processes.
The overall diversity of deep Arctic bacterial communities was lower than that detected by pyrosequencing in surface Arctic waters (Kirchman et al., Submitted), suggesting that Arctic bacterial communities from the dark ocean are less diverse than surface communities. We also detected half the observed number of OTUs and five times lower predicted richness (Chao1) on average than in an earlier pyrosequencing study from the deep North Atlantic (Sogin et al., 2006). The lower diversity of the Arctic may be explained by the latitudinal and temperature diversity gradient recently demonstrated for surface marine bacteria (Fuhrman et al., 2008), but the extent of the difference remains intriguing. Pyrosequencing richness was, however, more than 25 times greater than estimates from our own and other Arctic clone libraries (Malmstrom et al., 2007; Pommier et al., 2007), and molecular community fingerprinting studies (Fuhrman et al., 2008), consistent with hidden diversity unveiled using pyrosequencing and the tag approach (Sogin et al., 2006).
The bacterial communities of the deep Arctic Ocean were composed of groups previously defined as inhabitants of the meso- and bathypelagic water of the world's dark oceans. Among Alphaproteobacteria, we detected the deep water SAR11-D, earlier detected in deep waters of the Mediterranean Sea (Garcia-Martinez and Rodriguez-Valera, 2000) and Antarctic waters (Lopez-Garcia et al., 2001), thereby extending its record to the Arctic Ocean. The Deltaproteobacteria were dominated by the SAR324 cluster (Wright et al., 1997), another typical deep water cluster earlier reported from the Arctic (Bano and Hollibaugh, 2002) but also from the Antarctic (Lopez-Garcia et al., 2001) and North Atlantic (Gonzalez et al., 2000). The SAR406 cluster, previously detected in various oceanic provinces (Gordon and Giovannoni, 1996; Gallagher et al., 2004; Pham et al., 2008), as well as the Chloroflexi SAR202 cluster (Giovannoni et al., 1996), described as abundant and ubiquitous in meso- and bathypelagic waters (Wright et al., 1997; Bano and Hollibaugh, 2002; Morris et al., 2004; Varela et al., 2008a), were other abundant clusters of the dark ocean.
Within the abundant bacterial groups, we were able to identify OTUs specific to each water mass. Many OTUs were affiliated to uncultured organisms and the lack of metabolic information continues to hamper efforts to arrive at a functional characterization of different water masses. In some cases, however, the function of the bacteria may be inferred, for example, the most abundant group of Gammaproteobacteria (v6_CY086), common in the halocline of the Canada Basin, belonged to the GSO cluster of sulfide-oxidizing bacteria (Lavik et al., 2009).
Interestingly, patterns of biogeography were found within the classical deep-water clusters SAR406 and SAR202. Those clusters were composed of numerous subclusters with specific water mass distributions, which suggest that the ecological patterns of deep-water clusters are probably more complex than previously demonstrated. The present results also indicate that further quantitative studies on deep-water bacteria should focus on specific OTUs rather than on broader clusters detected with probes to fully understand the true ecological diversity of deep communities. In contrast, we note that such within cluster variability did not hold for all deep-water bacteria at the level of the 16S rRNA gene. For example, the highly abundant SAR324 cluster had low diversity and a ubiquitous distribution throughout the deep Arctic Ocean. It may indicate broader tolerance or adaptation of that group to the conditions common to many deep-water masses.
Our study revealed a strong association between the large-scale distribution of bacteria and the main water masses of the deep Arctic Ocean. Oceans are not uniform, but made up of regional water masses distinct in their temperature and salinity. Each water mass with its own environmental properties may carry a specific bacterial assemblage, and the water masses' history and local properties could explain bacterial biogeography. If such coupling between microbial and physical oceanography is valid at a global scale, it has to be carefully considered when studying the role and diversity of marine bacteria in the ocean.
Accession codes
References
Agogue H, Brink M, Dinasquet J, Herndl GJ . (2008). Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456: 788–791.
Azam F, Malfatti F . (2007). Microbial structuring of marine ecosystems. Nat Rev Micro 5: 782–791.
Bano N, Hollibaugh JT . (2002). Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol 68: 505–518.
Broecker WS . (1991). The great ocean conveyor. Oceanography 4: 79–89.
Broecker WS . (1997). Thermohaline circulation, the achilles heel of our climate system: will man-made CO2 upset the current balance? Science 278: 1582–1588.
Darling KF, Kucera M, Pudsey CJ, Wade CM . (2004). Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA 101: 7657–7662.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Dethlefsen L, Huse S, Sogin ML, Relman DA . (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Felsenstein J . (2004). PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author Department of Genome Sciences, University of Washington: Seattle, USA.
Field KG, Gordon D, Wright T, Rappe M, Urback E, Vergin K et al. (1997). Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol 63: 63–70.
Fuhrman JA, Davis AA . (1997). Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser 150: 275–285.
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci 105: 7774–7778.
Galand PE, Lovejoy C, Pouliot J, Garneau ME, Vincent WF . (2008). Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a Stamukhi lake and its source waters. Limnol Oceanogr 53: 813–823.
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C . (2009a). Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869.
Galand PE, Lovejoy C, Hamilton AK, Ingram RG, Pedneault E, Carmack E . (2009b). Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ Microbiol 11: 971–980.
Gallagher JM, Carton mW, Eardly DF, Patching JW . (2004). Spatio-temporal variability and diversity of water column prokaryotic communities in the eastern North Atlantic. FEMS Microbiol Ecol 47: 249–262.
Garcia-Martinez J, Rodriguez-Valera F . (2000). Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol Ecol 9: 935–948.
Giovannoni SJ, Rappe MS, Vergin KL, Adair NL . (1996). 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green Non-Sulfur bacteria. Proc Natl Acad Sci USA 93: 7979–7984.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782.
Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol 66: 4237–4246.
Gordon DA, Giovannoni SJ . (1996). Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol 62: 1171–1177.
Hamilton AK, Lovejoy C, Galand PE, Ingram RG . (2008). Water masses and biogeography of picoeukaryote assemblages in a cold hydrographically complex system. Limnol Oceanogr 53: 922–935.
Hammer Ø, Harper DAT, Ryan PD . (2001). PAST: Paleontological statistics software package for education and data analysis. Paleontol Electro 4: 9.
Hansman RL, Griffin S, Watson JT, Druffel ERM, Ingalls AE, Pearson A et al. (2009). The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA 106: 6513–6518.
Herndl GJ, Agogue H, Baltar F, Reinthaler T, Sintes E, Varela MM . (2008). Regulation of aquatic microbial processes: the ‘microbial loop’ of the sunlit surface waters and the dark ocean dissected. Aqua Microbial Ecol 53: 59–68.
Hewson I, Steele JA, Capone DG, Fuhrman JA . (2006). Remarkable heterogeneity in meso- and bathypelagic bacterioplankton assemblage composition. Limnol Oceanogr 51: 1274–1283.
Hosia A, Stemmann L, Youngbluth M . (2008). Distribution of net-collected planktonic cnidarians along the northern Mid-Atlantic Ridge and their associations with the main water masses. Deep-Sea Res II 55: 106–118.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huber T, Faulkner G, Hugenholtz P . (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319.
Huse S, Huber J, Morrison H, Sogin M, Welch D . (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143.
Jones EP, Rudels B, Anderson LG . (1995). Deep waters of the Arctic Ocean: origins and circulation. Deep Sea Res Pt I 42: 737–760.
Karl DM . (2002). Nutrient dynamics in the deep blue sea. Trends Microbiol 10: 410–418.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Konstantinidis KT, DeLong EF . (2008). Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J 2: 1052–1065.
Lavik G, Stuhrmann T, Bruchert V, Van der Plas A, Mohrholz V, Lam P et al. (2009). Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–584.
Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F . (2001). Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36: 193–202.
Macdonald RW, Carmack EC . (1991). Age of Canada Basin deep waters: a way to estimate primary production for the Arctic Ocean. Science 254: 1348–1350.
Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL . (2007). Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aqua Microbial Ecol 47: 45–55.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
McLaughlin FA, Carmack EC, Macdonald RW, Melling H, Swift JH, Wheeler PA et al. (2004). The joint roles of Pacific and Atlantic-origin waters in the Canada Basin, 1997–1998. Deep Sea Res Pt I 51: 107–128.
Morris RM, Rappe MS, Urbach E, Connon SA, Giovannoni SJ . (2004). Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol 70: 2836–2842.
Pham VD, Konstantinidis KT, Palden T, DeLong EF . (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol 10: 2313–2330.
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Rudels B, Jones PE, Schauer U, Eriksson P . (2004). Atlantic sources of the Arctic Ocean surface and halocline waters. Polar Res 23: 181–208.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Sexton PF, Norris RD . (2008). Dispersal and biogeography of marine plankton: long-distance dispersal of the foraminifer Truncorotalia truncatulinoides. Geology 36: 899–902.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the under explored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Spencer-Cervato C, Thierstein H . (1997). First appearance of Globorotalia truncatulinoides: Cladogenesis and immigration. Mar Micropaleontol 30: 267–291.
Stevens H, Ulloa O . (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol 10: 1244–1259.
Stommel H . (1958). The abyssal circulation. Deep-Sea Res 5: 80–82.
Suzuki MT, Béjà O, Taylor LT, DeLong EF . (2001). Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol 3: 323–331.
Tang CCL, Ross CK, Yao T, Petrie B, DeTracey BM, Dunlap E . (2004). The circulation, water masses and sea-ice of Baffin Bay. Prog Oceanogr 63: 183–228.
Teira E, Lebaron P, van Aken H, Herndl GJ . (2006). Distribution and activity of bacteria and archaea in the deep water masses of the North Atlantic. Limnol Oceanogr 51: 2131–2144.
Varela MM, van Aken HM, Herndl GJ . (2008a). Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ Microbiol 10: 1903–1911.
Varela MM, van Aken HM, Sintes E, Herndl GJ . (2008b). Latitudinal trends of Crenarchaeota and bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ Microbiol 10: 110–124.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Wright TD, Vergin KL, Boyd PW, Giovannoni SJ . (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448.
Acknowledgements
We deeply appreciate the assistance and support of the men and women of the Canadian Coast Guard icebreaker CCGS Louis St Laurent, and we acknowledge financial and ship time support from Fisheries and Oceans Canada and the Canadian International Polar Year Program's Canada's Three Oceans project and the Nansen and Amundsen Basins Observational System project. PE Galand was supported by a Marie Curie Grant (CRENARC MEIF-CT-2007-040247) and EO Casamayor by the Spanish Grant CGL2006-12058-BOS. C Lovejoy would like to acknowledge the support of the Natural Sciences and Engineering Council, Canada (NSERC) Special Research Opportunity Fund and ArcticNet. Deep Arctic samples were collected by K Scarcella, E Didierjean and M-É Garneau. Pyrosequencing was supported by a Keck foundation grant to M Sogin and L Ameral Zettler. This is a contribution to the International Census of Marine Microbes (ICOMM). We also thank three anonymous reviewers for suggestions and constructive comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary information
Rights and permissions
About this article
Cite this article
Galand, P., Potvin, M., Casamayor, E. et al. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J 4, 564–576 (2010). https://doi.org/10.1038/ismej.2009.134
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2009.134
Keywords
This article is cited by
-
The distribution and diversity of eukaryotic phytoplankton in the Icelandic marine environment
Scientific Reports (2023)
-
Vertical segregation and phylogenetic characterization of archaea and archaeal ammonia monooxygenase gene in the water column of the western Arctic Ocean
Extremophiles (2023)
-
Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges
Scientific Reports (2022)
-
Major ocean currents may shape the microbiome of the topshell Phorcus sauciatus in the NE Atlantic Ocean
Scientific Reports (2021)
-
Comparison of picoeukaryote community structures and their environmental relationships between summer and autumn in the southern Chukchi Sea
Extremophiles (2021)