Abstract
The first backcross breeding programme for the transfer of freezing-tolerance genes from winter hardy Festuca pratensis to winter-sensitive Lolium multiflorum is described. A partly fertile, triploid F1 hybrid F. pratensis (2n=2x=14) × L. multiflorum (2n=4x=28) was employed initially, and after two backcrosses to L. multiflorum (2x) a total of 242 backcross two (BC2) plants were generated. Genomic in situ hybridisation (GISH) was performed on 61 BC2 plants selected for their good growth and winter survival characters in the spring following one Polish winter (2000–2001). Among the winter survivors, diploid chromosome numbers were present in 80% of plants. An appropriate single Festuca introgression in an otherwise undisturbed Lolium genome could provide increased freezing tolerance without compromise to the good growth and plant vigour found in Lolium. Among all the diploids, a total of 20 individuals were identified, each with a single F. pratensis chromosome segment. Another diploid plant contained 13 Lolium chromosomes and a large metacentric F. pratensis chromosome, identified as chromosome 4, with two large distal Lolium introgressions on each chromosome arm. Three of the diploid BC2, including the genotype with Festuca chromosome 4 DNA sequences, were found to have freezing tolerance in excess of that of L. multiflorum, and in one case in excess of the F. pratensis used as control. A detailed cytological analysis combining GISH and fluorescence in situ hybridisation analyses with rDNA probes revealed that the other two freezing-tolerant genotypes carried a Festuca chromosome segment at the same terminal location on the non-satellite arm of Lolium chromosome 2.
Similar content being viewed by others
Introduction
The species within the Lolium–Festuca complex comprise a range of desirable and complementary characters. Lolium multiflorum (2n=2x=14; Lm) and L. perenne (2n=2x=14; Lp) are the most important forage grasses for European agriculture, with high productivity and quality but rather poor persistency under stress conditions. Conversely, although having relatively inferior seedling vigour and nutritive value, closely related Festuca pratensis (2n=2x=14; Fp), F. glaucescens (2n=4x=28; Fg), and F. arundinacea (2n=6x=42; Fa) can provide a greater tolerance to climatic stresses than Lolium. These Lolium and Festuca species hybridise readily and their homoeologous chromosomes pair and recombine at high frequency, enabling the assembly of their complementary characters within a single genotype (Humphreys et al, 1997). Intergeneric amphiploids that combine complementary characters of both genera have been generated (eg Lewis et al, 1973; Zwierzykowski et al, 1998; Casler et al, 2001), but homoeologous chromosome pairing has led often to aneuploidy, genetic imbalance, and sterility in the hybrids, limiting breeding progress. Furthermore, a shift towards the Lolium parental genotype in amphiploid L. perenne or L. multiflorum × F. pratensis over several generations (up to F8) has been observed by different research groups (Zwierzykowski et al, 1998, 2003; Canter et al, 1999; Pašakinskienė and Jones, 2003). As recombination between homoeologous Lolium and Festuca chromosomes occurs frequently in hybrids, opportunities for gene introgression arise, enabling the transfer to Lolium of Festuca genes for improved persistency, through a backcross breeding programme (Thomas et al, 1994, 2003; Humphreys et al, 1997, 2003, 2005; King et al, 1998; Zwierzykowski et al, 1998, 1999). The targeted inclusion of desirable Festuca gene combinations can be accompanied by the targeted exclusion of other Festuca genes considered detrimental to the high forage quality traits found in Lolium.
Molecular cytogenetics has not only revolutionised the genetic analysis of plant genomes, but has also provided plant breeders with new tools to identify genes involved in resistance to abiotic and biotic stresses. The development of the genomic in situ hybridisation (GISH) technique for cytogenetic analyses in Lolium–Festuca hybrids has represented a major advance in the genome analysis of these genera (eg Bailey et al, 1993; Thomas et al, 1994; Humphreys and Pašakinskienė, 1996; Zwierzykowski et al, 1998; Leśniewska et al, 2001; Pašakinskienė and Jones, 2005). GISH enables the parental chromosomes of Lolium and Festuca to be distinguished, and the sites of any genome recombination identified. GISH also enables the monitoring of the introgression of alien chromatin from one species to another in different generations during backcross breeding programmes (eg Humphreys and Pašakinskienė, 1996; Zwierzykowski et al, 1998, 1999).
One of the limiting factors for the successful widespread use of high-yielding Lolium cultivars is their susceptibility to winter stresses. As a consequence, the combining of the nutritive quality of L. multiflorum with the winter hardiness of F. pratensis together in a single genotype is considered a primary grass breeding objective (Humphreys et al, 1998).
Jones et al (2002) performed the first comparative mapping study between forage grasses and cereals by comparing L. perenne with the Triticeae, oats, and rice. Recently, Inoue et al (2004) and Sim et al (2005) in Lolium species and Alm et al (2003) in F. pratensis have extended the comparison further. They found that the genetic maps of Lolium and Festuca species and the Triticeae cereals were highly conserved in terms of orthology (chromosomal segment structure) and colinearity (marker order). Moreover, there was considerable synteny between L. perenne, L. multiflorum and F. pratensis. Lolium and Festuca spp chromosomes and linkage groups described herein are thus numbered in accordance with their orthologous Triticeae chromosome counterparts.
The work presented in this paper demonstrates for the first time how a backcross breeding programme enabled the transfer of F. pratensis genes for freezing tolerance into diploid freezing-sensitive L. multiflorum cultivars using fertile, triploid F. pratensis (2n=2x=14) × L. multiflorum (2n=4x=28) hybrids. The approach was similar to that described previously by Grønnerød et al (2004), who used an amphiploid L. perenne × F. pratensis cultivar as a starting point for the introgression of F. pratensis genes for freezing tolerance into L. perenne. However, the regions of the genome targeted as sources for genes for freezing tolerance differ in the current study. The approach described herein includes the cytogenetic mapping of putative genes for freezing resistance using GISH in association with a fluorescence in situ hybridisation (FISH) study employing rDNA probes to assist with chromosome identification. The targeting of diploid (2n=2x=14) freezing-tolerant introgression forms of L. multiflorum that contain only one F. pratensis chromosome segment allowed the location of putative Festuca-derived genes for freezing tolerance. The main objectives of our research were (1) to screen the genomic structure of backcross 2 (BC2) plants and to identify diploid L. multiflorum (Lm) genotypes with a single F. pratensis (Fp) chromosome segment, (2) to test if Fp genes transferred to Lm led to an improvement in freezing tolerance, and (3) to identify the chromosome location of putative Fp genes for freezing tolerance introgressed into Lolium.
Materials and methods
Plant material
The intergeneric hybrids for the backcross programme were produced by reciprocally crossing autotetraploids (2n=4x=28) L. multiflorum Lam. (cvs. Atos, Lotos, Gran) (designated LmLmLmLm) with diploids (2n=2x=14) F. pratensis Huds. (cvs. Skrzeszowicka, Merbeen, Lifesta) (designated FpFp). Seven partially fertile triploids (2n=3x=21) including genotype 45/4/29 LmLmFp (with Lm cytoplasm) and six genotypes, 57/1/17, 57/1/28, 57/3/1, 57/3/4, 57/3/6, 57/6/2 FpLmLm (with Fp cytoplasm), were backcrossed as male parents onto seven different genotypes of diploid Lolium cultivars Adret, Abercomo, and Tur (designated LmLm). As a result of these backcrosses, seven backcross 1 (BC1) populations (nos. 60/6, 66/2, 66/4, 66/10, 66/8, 66/7, 66/11) and a total of 154 BC1 plants were generated. These BC1 plants were reported by Zwierzykowski et al (1999) to range in chromosome number between 14 and 23, and were the progenitors of the plant generation described in this paper. A total of 14 plants derived from all the BC1 populations, selected on the basis of their high vigour and good fertility, were backcrossed again (as a male or as a female parent) to diploid Lm cvs. Abercomo, Adret, and Tur to produce 12 BC2 populations. After initial establishment in the glasshouse, BC2 plants were transferred to the field and subsequently selected in the spring following the winter of 2000/2001 for winter hardiness. Winter survivors with the highest plant vigour were chosen for cytogenetic analyses. Figure 1 provides a schematic diagram of the chronological order of events described in the current study.
A schematic diagram of the breeding programme, plant selection, and physical mapping with GISH and FISH of F. pratensis (Fp) genes for freezing tolerance transferred from Festuca to the diploid L. multiflorum genome. The BC2 plants were generated by reciprocal crosses involving BC1 plants as either female or male parents in backcrosses with L. multiflorum (2x).
Cytogenetic analyses
To determine the chromosome number and genomic structure of BC2 plants, root-tip chromosome spreads were prepared as described by Zwierzykowski et al (1999). Chromosome preparations were dried at room temperature and stored at −20°C.
Genomic in situ hybridisation
To discriminate between Lm and Fp chromosomes, total genomic DNA of Lm cv. Tur was used as a probe and labelled with digoxigenin-11-dUTP by nick translation (according to the Roche protocol). Total genomic DNA of Fp cv. Skra to be used as a blocker was prepared by autoclaving for 10 min and applied at a ratio of 1:40 (probe:blocker). The GISH protocol was that of Zwierzykowski et al (1998) with one minor modification: the slides were incubated with anti-digoxigenin-fluorescein (Roche) at the final concentration of 2 μg/ml at 37°C for 40 min. Preparations were analysed using a Nikon Optiphot-2 epifluorescence microscope and photographed on Fuji 800 films. For each plant, the total number of chromosomes, the number of chromosomes from each parent, and the number of introgressions involving the transfer of Lm genes onto Fp chromosomes or Fp genes onto Lm chromosomes were determined. Only diploid plants (2n=2x=14) carrying a single introgressed Fp chromosome segment were used in the freezing tests described below. Chromosome preparations of these diploid introgression lines were re-probed using rDNA probes in FISH experiments.
Fluorescence in situ hybridisation
To detect 45S rRNA genes, an 18S–25S rDNA fragment from Arabidopsis thaliana in the plasmid SK+ was labelled with digoxigenin-11-dUTP by nick translation (protocol according to the manufacturer Roche). To detect 5S rRNA genes, a plasmid clone pU5SLL, which contained one unit of 5S rDNA from Lupinus luteus L., was also used (Nuc et al, 1993). DNA was labelled with biotin-16-dUTP in a standard PCR using 20-mer primers corresponding to the 5′ and 3′ ends of the 119-bp-long coding region, without the non-transcribed spacer. After GISH, the coverslips were removed and the slides were washed twice in 2 × SSC at room temperature for 5 min, followed by three 45-min washes in 4 × SSC/0.05% Tween 20 at 60°C. From the step of chromosome denaturation in 70% formamide/2 × SSC, FISH protocols were as described for GISH. The hybridisation mixture consisted of 100 ng of each probe, 10 μg herring sperm DNA, 10% dextran sulphate, 50% formamide, 2 × SSC, and 0.25% SDS. For 5S and 45S rDNA detection, a one-step procedure was used and the slides were incubated with anti-digoxigenin-fluorescein (Roche) and Cy3-conjugated streptavidin (Sigma), both at a final concentration of 1 μg/ml at 37°C for 30 min. The slides were mounted in Vectashield (Vector Laboratories, Burlingame, CA, USA) with 0.2 μg DAPI dihydrochloride/ml as a counterstain. The preparations were examined using an Olympus BX-60 microscope (Olympus Optical Co., GmBH, Hamburg, Germany). An uncooled charge-coupled device camera, Ikegami 47E, was used and images were processed by the AnalySIS 3.0 program (SIS® Soft Imaging System GMBH, Münster, Germany). For karyotyping, the images of the same mitotic chromosome spreads after GISH and FISH were compared and thereby the identity of the Lm chromosomes carrying Fp introgressions was determined.
Test for freezing tolerance
Before cold acclimation, seedlings of BC2 plants were divided into 12 equal-sized clones of five tillers per genotype, and planted into three boxes each containing a sand:peat (1:1) mixture. Four replicate clones of each genotype were planted randomly in each of the three boxes to be used in the freezing test. As control, plants of four different genotypes of Lm cv. Tur and Fp cv. Fure were used. Each genotype was divided into three equal-sized clones (one clone per each of the three boxes). Plants were established in the sand/peat mixture over 7 days (25°C, 10/14 h photoperiod day/night, and 200 μmol/m2 s photosynthetic photon flux density (PPFD), Philips AGRO sodium light source, Philips Lightning NV, Turnhout, Belgium). Well-rooted plants were then transferred to a controlled environment (CE) chamber for pre-hardening (7 days at 12°C, 8/16 h photoperiod, and 200 μmol/m2 s PPFD) and then for cold acclimation (3 weeks at 2°C, 10/14 h photoperiod, and 200 μmol/m2 s PPFD). Plant freezing tolerance was determined by a modified method of that described by Rapacz et al (2004). Cold-acclimated plants were transferred for 24 h to a CE chamber at –2°C, 10/14 h photoperiod, and 200 μmol/m2 s PPFD and then for 20 h in the dark at –4°C. After this time, plants were frozen at a cooling rate of approximately 1°C/h to the desired freezing temperature: box 1 to –7°C, box 2 to –11°C, and box 3 to –15°C. Following an 8-h exposure to the target temperature, each box was transferred to 2°C, 10/14 h photoperiod, and 200 μmol/m2 s PPFD to defrost, and 24 h later, plants were transferred to the CE chamber at 12°C, 12/12 h photoperiod, and 300 μmol/m2 s PPFD, to recover over 23 days, which is normally sufficient for the re-growth of tillers (Larsen, 1978). All plants were cut down to a height of 2–3 cm after hardening, and the re-growth of four clones of each genotype per particular freezing temperature was estimated using Larsen's (1978) visual score. This was as follows: 0=dead, no sign of leaf elongation; 1=dead, but leaves having previously elongated to approximately 5 mm; 2=dead, but leaves having previously elongated to 1–2 cm; 3=plant dying, but with leaves having elongated to >2 cm; 4=plant likely to die and with inner leaves brown; 5=plant likely to survive, but badly damaged; 6=plant surviving, but with severe damage to approximately 50% of the leaves; 7=plant alive, but with visual signs of freezing injury; 8=minimal freezing damage (leaf tips discoloured or deformed); 9=no visible sign of injury. Mean scores for four clones of each plant following recovery were determined, and the mean scores for each genotype at all the three freezing temperatures were calculated. Statistical significance for differences between plants in freezing tolerance was determined using Duncan's multiple range test at P=0.05.
Results
Development of BC2 populations
A total of 2460 Lm and BC1 florets (Lm total=1413; BC1 total=1047) were pollinated in 12 reciprocal cross-combinations. The overall mean seed set was 21%, but seed set differed greatly between populations ranging between 2.9% (population 95/15) and 69.3% (population 95/1). The values were dependent on whether the Lm or BC1 individual was used as the female parent. When Lm was used as the female parent, the mean seed set was 30%, but when the BC1 plant was used as the female parent, the mean seed set was only 11% (Table 1). Seedling vigour was also higher when Lm was employed as the female parent. Among a total of 242 BC2 plants established before the winter field trial, only 54 (22%) arose from crosses involving the BC1 plant as the female parent. Following the winter field trial, a total of 129 (53.3%) survivors were recovered, of which 95 were plants derived from crosses where Lm was employed as the female parent and 34 were plants where the BC1 plant was used as the female parent (Table 1).
Genomic structure of BC2 plants
A total of 61 BC2 plants (derived from 11 cross-combinations) with the highest plant vigour following the winter survival field tests were analysed cytologically (Table 2). The chromosome number in these plants ranged from 14 to 18, but the diploid chromosome number predominated, especially when Lm was used as the female parent. Only three aneuploids (7%) were recovered among a total of 42 BC2 genotypes in populations derived from an Lm female parent. This contrasted with nine aneuploids (47%) among a total of 19 BC2 genotypes in populations derived when the BC1 plant was employed as the female parent. The numbers of introgressions also differed, depending on the direction of the cross, and were higher when the BC1 plant was used as the female parent. The frequencies of recovery of Fp introgressions on Lm chromosomes were the same, irrespective of whether Lm or the BC1 plants were used as female parents. However, their numbers were greater than Lm introgressions on Fp chromosomes irrespective of whether Lm or BC1 plants were used as female parents. Among the 19 BC2 genotypes derived from a BC1 female parent plant, a total of 62 introgressions were detected of which 42 (68%) were Fp introgressions on Lm chromosomes. In contrast, among the 42 BC2 genotypes derived from an Lm female parent, a total of 58 introgressions were observed of which 41 (71%) were Fp introgressions on Lm chromosomes. Only six (12%) from a total of 49 diploid BC2 genotypes carried no observable introgressed chromosome segment. In population 95/9, all 16 plants were diploids with 11 plants carrying a single Fp introgression, each located terminally on a large Lm chromosome.
Among all the BC2 plants with the euploid 14 chromosome number, examples were found of genotypes with one or two Fp chromosomes, each having replaced its Lm homoeologue. One diploid BC2 plant (no. 95/12/24) had 13 Lm chromosomes and a chromosome with an Fp-derived centromere and Fp-derived pericentromeric regions, as well as two large distal Lm translocations on each chromosome arm.
Test for freezing tolerance
Six BC2 plants were chosen for a freezing-tolerance test on the basis of the GISH analysis (each having a single Fp introgression) and of having sufficient size to be divided into the required numbers of replicates. These were 95/9/2, 95/9/6, 95/9/15, 95/9/18, 95/9/29, and 95/12/24. They varied greatly in their freezing tolerance. Mean freezing-tolerance scores for four clones of each BC2 genotype, and for Lm Tur and Fp Fure controls following 23 days recovery following freezing, are shown in Table 3. A Duncan's multiple range test indicated that Lm Tur and BC2 introgression forms 95/9/2, 95/9/15, and 95/9/18 all grouped together as freezing-sensitive genotypes. All the other plants were significantly (P=0.05) better with respect to freezing tolerance. Some suggestion of transgressive segregation was observed in plant no. 95/9/29, which was significantly more freezing tolerant than the Fp Fure control. Among the six BC2 plants examined for freezing tolerance, three plants (nos. 95/9/6, 95/9/29, and 95/12/24) were significantly more tolerant than Lm.
Detailed GISH/FISH cytological analysis of elite freezing-tolerant genotypes
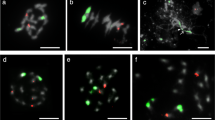
The chromosome morphology of freezing-tolerant BC2 genotypes 95/9/6, 95/9/29, and 95/12/24 was studied in detail. FISH analysis using rDNA probes aids the identification of the three Lm chromosomes 2, 3, and 7 with primary and secondary constrictions (MW Humphreys, unpublished). For each plant cell analysed, six signals for the 45S rDNA probe on secondary constructions of homologous pairs of chromosomes 2, 3, and 7 and two signals for 5S rDNA probe on a pair of homologues of chromosome 2 were observed (Figure 2b, d, and f). After comparing GISH and FISH images of the same mitotic chromosome spreads, it was revealed that in the case of both 95/9/6 (Figure 2a and b) and 95/9/29 (Figure 2c and d), a Fp segment was located terminally on the short non-satellite arm of chromosome 2 (chromosome with both 45S and 5S rDNA loci).
GISH (a, c, e) and FISH (b, d, f) analyses of the same mitotic chromosome spreads of freezing-tolerant plants: BC2-95/9/6 (a, b), 95/9/29 (c, d), and 95/12/24 (e, f). GISH images (a, c, e) created using total genomic DNA of L. multiflorum as a probe labelled with digoxigenin and detected by anti-digoxigenin conjugated with fluorescein (yellow), with blocking genomic DNA of F. pratensis (red). Chromosomes were counterstained with propidium iodide. FISH images (b, d, f) created using as a probe (i) 5S rDNA labelled with biotin and detected by streptavidin conjugated with Cy-3 (red) and (ii) 45S rDNA labelled with digoxigenin and detected by anti-digoxigenin conjugated with fluorescein (green). Chromosomes for FISH analysis were counterstained with DAPI. GISH (a, c) and FISH (b, d) analyses indicate that BC2-95/9/6 and BC2 95/9/29 carry F. pratensis introgressions on the non-satellite arm of chromosome 2 of Lolium. No rDNA sites were detected (f) on the BC2 95/12/24 chromosome containing the F. pratensis sequences, identified as chromosome 4.
The large median Fp chromosome in the freezing-tolerant plant 95/12/24, which contained Lm introgressions on both chromosome arms, carried no rDNA sites. The 45S and 5S rDNA loci were detected elsewhere in the genome (Figure 2e and f). The Fp chromosome was identified as chromosome 4 based on its large size and the median location of its centromere.
Discussion
Introgression in the Lolium/Festuca complex using triploid F1 intergeneric hybrids as male parents in backcrosses onto diploid recurrent Lolium parents has been demonstrated previously to work very effectively with the diploid Lolium chromosome complement recovered in 1–2 generations (Thomas et al, 1988, 2003; Humphreys et al, 2003). The use of the F1 hybrid and backcross progeny as the male parent in each backcross accelerates the loss of chromosomes of the non-recurrent parent because pollen with a near or complete haploid Lolium genome has a selective advantage (Morgan et al, 1988; Thomas et al, 1988; Humphreys et al, 1998). Diploid hybrids of Lolium spp and F. pratensis are generally male and female sterile (Jauhar, 1975). Triploid hybrids with a diploid set of Lolium chromosomes and a haploid set of Festuca chromosomes have considerable pollen fertility. However, those with a diploid set of Festuca chromosomes and a haploid set of Lolium chromosomes remain infertile (Jauhar, 1975; Naganowska et al, 2001; Thomas et al, 2003). Herein, it was shown that the choice of Lolium or BC1 as female parents also had a significant effect on the numbers of diploid BC2 genotypes created, the numbers of intergeneric recombinants recovered, and on plant vigour and its capacity for winter survival. After using an L. perenne (4x) × F. pratensis (2x) hybrid as the male parent in backcrosses to diploid L. perenne, King et al (1998) reported that 96.2% plants of BC1 progeny had 14 chromosomes. Moreover, 77% diploid plants carried only one, two, or three Fp chromosome segments. Conversely, Zwierzykowski et al (1999), using a triploid L. multiflorum (4x) × F. pratensis (2x) hybrid as the female parent, revealed that only 51.5% of BC1 progeny were diploids.
In the current study, more of the Festuca genome was retained in the BC2 plant when the BC1 plant was used as the female parent than in the reciprocal cross, but this led to an overall reduction in seed set and a reduction in seedling survival. However, irrespective of the direction of the cross, a similar imbalance was found between numbers of Fp introgressions on Lm chromosomes and Lm introgressions on Fp chromosomes. In reciprocal crosses, Fp introgressions on Lm chromosomes exceeded Lm introgressions recovered on Fp chromosomes. During meiosis, intergeneric chromosome pairing and numbers of resulting recombinants involving Lm and Fp chromosomes must initially be equal. Thus, there must be subsequently either preferential selection for Fp introgressions on Lm chromosomes or selection against Lm introgressions present on Fp chromosomes, or simply selection against the presence of Fp chromosomes. Irrespective of the reduced numbers of Fp introgressions in the backcross populations developed from crosses involving the Lm as female, the majority of field survivors following a Polish winter originated from these populations. Furthermore, the most freezing-tolerant genotype 95/9/29, which exceeded the tolerance found in both Lm and Fp control genotypes, was developed from a cross involving Lm as the female parent. The three freezing-tolerant introgression lines selected all carried only a single Fp introgression and were all significantly more tolerant (P=0.05) than the Lm control. This is the first report of the transfer of genes for freezing tolerance between Fp and Lm and follows a similar success in the transfer of Fp genes for freezing tolerance into L. perenne (Grønnerød et al, 2004), although in that case genes on chromosome 3 of Fp were involved. However, the significance of the Fp introgressions reported here is greater, as Lm is more sensitive to freezing conditions than L. perenne and the benefits to accrue from the Fp introgressions reported herein are therefore more significant. Despite freezing tolerance being a quantitative character governed by many genes, it is demonstrated here, as was reported previously for drought resistance (Humphreys and Pašakinskienė, 1996; Humphreys et al, 2005), that a single Festuca introgression can make a significant impact on the resistance of Lolium against abiotic stresses.
The application of GISH/FISH techniques in the physical mapping of abiotic stress resistance genes in different introgression lines derived from triploid, tetraploid, and pentaploid intergeneric hybrids has been reported previously. For example, in diploid L. multiflorum introgression forms obtained from the backcrossing of L. multiflorum (4x) × F. glaucescens (4x) hybrid, a Festuca introgressed segment with genes governing drought resistance has been located on the NOR arm of Lolium chromosome 3 (Humphreys et al, 2005). In this study, clones of pTa/71 containing a fragment of 18S–5.8S–26S rDNA from Triticum aestivum and pTa794 (with a fragment of 5S rDNA from T. aestivum) were used for chromosome identification. These clones had been applied previously for karyotyping of Lolium and Festuca species (Thomas et al, 1996, 1997). In the research described herein, DNA fragments from dicotyledonous species A. thaliana and L. luteus (Nuc et al, 1993) were used to prepare 45S and 5S rDNA probes, respectively. It was shown previously that the coding regions of 45S and 5S rDNA were strongly conserved even between distantly related species (eg Arnheim, 1983; Hillis and Dixon, 1991; Allaby and Brown, 2001; Volkov et al, 2003). These genes have been used elsewhere as probes in heterologues hybridisation (eg Hasterok et al, 2001; Ziółkowski and Sadowski, 2002).
Recent information concerning the location of genes for freezing tolerance in F. pratensis has been derived from a QTL mapping approach (Alm et al, 2006). They have been located in linkage groups 4–7. The introgression mapping approach described here indicates that regions of chromosomes 2 and 4 are involved in both winter hardiness and freezing tolerance. A number of genes of importance that are implicated in winter hardiness have been identified on these chromosomes in the Triticeae. Recently, the vernalisation locus Vrn1 mapped onto wheat chromosome 5A (Galiba et al, 1995) was located on chromosome 4 in L. perenne (Jensen et al, 2005). Moreover, on the same chromosome in perennial ryegrass, close to a heading date locus, was identified a QTL for electrical conductivity corresponding to frost tolerance (Yamada et al, 2004). Principal genes for photoperiod response, which could also be involved in the regulation of freezing tolerance, have been mapped on wheat chromosome 2 (Law et al, 1978; Scarth and Law, 1983, 1984; Sourdille et al, 2000). It remains to be found whether these or alternative Fp-derived genes are responsible for generating the enhanced freezing tolerance now observed in Lm following their successful transfer from Fp. The selection among the freezing-tolerant genotypes of two BC2 plants 95/9/6 and 95/9/29, both carrying the same Fp-derived introgression on chromosome 2, makes further study of this chromosome region a priority. We believe that the three ‘elite’ freezing-tolerant plants described here will be a good starting point for investigations in comparative genomics and proteomics, and for the transfer of biochemical, genetic, and physiological information from well-studied cereal species to Fp. A third backcross of the selected freezing-tolerant BC2 plants into diploid cultivars of Lm has been performed and will include genotypes where the Fp introgressions have been further ‘dissected’ through chromosome recombination thereby becoming the basis for our further analysis.
Conclusions
Described herein for the first time were procedures using fertile triploid F. pratensis (2n=2x=14) × L. multiflorum (2n=4x=28) hybrids as the starting point for the successful transfer of winter hardiness and freezing-tolerance traits from F. pratensis to L. multiflorum during a backcross breeding programme. The use of the BC1 plant as male parent led to the recovery of greater numbers of diploid Lolium-like BC2 plants than when it was used as the female parent. Despite the greater numbers of Festuca genes recovered among the BC2 populations when the BC1 plant was used as the female parent, this gave no obvious advantage to winter survival. An appropriate single Festuca introgression in an otherwise undisturbed Lolium genome provided increased freezing tolerance without compromise to the good growth and vigour found in Lolium. The combined use of GISH and FISH with two rDNA probes enabled the chromosome location of putative Festuca genes for freezing tolerance on chromosomes 2 and 4 that subsequent to their transfer from Festuca had led to enhanced freezing tolerance of Lolium. These chromosomes in the Triticeae are known to contain genes involved in both winter hardiness and freezing tolerance. A comparative genomics study will determine whether major genes concerned with these traits are conserved across other monocot crop species.
References
Allaby RG, Brown TA (2001). Network analysis provides insights into evolution of 5S rDNA arrays in Triticum and Aegilops. Genetics 157: 1331–1341.
Alm V, Busso CS, Larsen A, Humphreys MW, Rognli OA (2006). Quantitative trait loci for frost tolerance, winter survival and drought tolerance in meadow fescue (Festuca pratensis Huds.), and comparative mapping within cereals. Genetics (in press).
Alm V, Fang C, Busso CS, Devos KM, Vollan K, Grieg Z et al (2003). A linkage map of meadow fescue (Festuca pratensis Huds.) and comparative mapping with the Triticeae species, Lolium, oat, rice, maize and sorghum. Theor Appl Genet 108: 25–48.
Arnheim N (1983). Concerted evolution of multigene families. In: Nei M, Koehn RK (eds) Evolution of Genes and Proteins. Sinauer Associates: Sunderland, MA. pp 38–61.
Bailey JP, Bennett ST, Bennett MD, Stace CA (1993). Genomic in situ hybridization identifies parental chromosomes in the wild grass hybrid × Festulpia hubbardii. Heredity 71: 413–420.
Canter PH, Pašakinskienė I, Jones RN, Humphreys MW (1999). Chromosome substitution and recombination in the amphiploid Lolium perenne × Festuca pratensis cv Prior (2n=4x=28). Theor Appl Genet 98: 809–814.
Casler MD, Pitts PG, Rose-Ficker C, Bilkey PC, Wipff JK (2001). Registration of ‘Spring Green’ Festulolium. Crop Sci 41: 1365–1366.
Galiba G, Quarrie SA, Sutka J, Morgounov A, Snape JW (1995). RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet 90: 1174–1179.
Grønnerød S, Fjellheim S, Humphreys MW, Østrem L, Canter PH, Grieg Z et al (2004). Application of AFLP and GISH techniques for identification of Festuca chromosome segments conferring winter hardiness in a Lolium perenne × Festuca pratensis population. In: Hopkins A, Wang ZY, Mian R, Sladge M, Backer RE (eds) Molecular Breeding of Forage and Turf. Kluwer Academic Publishers: The Netherlands. pp 81–86.
Hasterok R, Jenkins G, Langdon T, Jones RN, Małuszyńska J (2001). Ribosomal DNA is an effective marker of Brassica chromosomes. Theor Appl Genet 103: 486–490.
Hillis DM, Dixon MT (1991). Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 66: 411–453.
Humphreys J, Harper JA, Armstead IP, Humphreys MW (2005). Introgression-mapping of genes for drought resistance transferred from Festuca arundinacea var. glaucescens into Lolium multiflorum. Theor Appl Genet 110: 579–587.
Humphreys MW, Canter P, Thomas HM (2003). Advances in introgression technologies for precision breeding within the Lolium–Festuca complex. Ann Appl Biol 143: 1–10.
Humphreys MW, Pašakinskienė I (1996). Chromosome painting to locate genes for drought resistance transferred from Festuca arundinacea to Lolium multiflorum. Heredity 77: 530–534.
Humphreys MW, Pašakinskienė I, James AR, Thomas H (1998). Physically mapping quantitative traits for stress-resistance in the forage grasses. J Exp Bot 49: 1611–1618.
Humphreys MW, Thomas HM, Harper J, Morgan M, James A, Zare AG et al (1997). Dissecting drought- and cold-tolerance traits in the Lolium–Festuca complex by introgression mapping. New Phytol 137: 55–60.
Inoue M, Gao Z, Hirata M, Fujimori M, Cai H (2004). Construction of a high-density linkage map of Italian ryegrass (Lolium multiflorum Lam.), using restriction fragment length polymorphism, amplified fragment length polymorphism, and telomeric repeat associated sequence markers. Genome 47: 57–65.
Jauhar PP (1975). Chromosome relationships between Lolium and Festuca (Gramineae). Chromosoma 52: 103–121.
Jensen LB, Andersen JR, Frei U, Xing Y, Taylor C, Holm PB et al (2005). QTL mapping of vernalization response in perennial ryegrass (Lolium perenne L.) reveals co-location with an orthologue of wheat VRN1. Theor Appl Genet 110: 527–536.
Jones ES, Mahoney NL, Hayward MD, Armstead IP, Jones JG, Humphreys MO et al (2002). An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome 45: 282–295.
King IP, Morgan WG, Armstead IP, Harper JA, Hayward MD, Bollard A et al (1998). Introgression mapping in the grasses. I. Introgression of Festuca pratensis chromosomes and chromosome segments into Lolium perenne. Heredity 81: 462–467.
Larsen A (1978). Freezing tolerance in grasses. Methods for testing in controlled environments. Department of Farm Crops Report No. 195, Sci Rep Agr Univ Norway.
Law CN, Sutka J, Worland AJ (1978). A genetic study of day-length response in wheat. Heredity 41: 185–191.
Leśniewska A, Ponitka A, Ślusarkiewicz-Jarzina A, Zwierzykowska E, Zwierzykowski Z, James AR et al (2001). Androgenesis from Festuca pratensis × Lolium multiflorum amphidiploid cultivars in order to select and stabilise rare gene combinations for grass breeding. Heredity 86: 167–176.
Lewis EJ, Tyler BF, Charlton KH (1973). Development of Lolium–Festuca hybrids. Report of the Welsh Plant Breed Station for 1972, pp 34–37.
Morgan WG, Thomas H, Lewis EJ (1988). Cytogenetic studies of hybrids between Festuca gigantea Vill. and Lolium multiflorum Lam. Plant Breeding 118: 491–496.
Naganowska B, Zwierzykowski Z, Zwierzykowska E (2001). Meiosis and fertility of reciprocal hybrids of Lolium multiflorum with Festuca pratensis. J Appl Genet 42: 247–255.
Nuc KT, Nuc PW, Pawełkiewicz J (1993). The nucleotide sequence and organization of nuclear 5S rRNA genes in yellow lupine. Bull Pol Acad Sci Chem 41: 103–106.
Pašakinskienė I, Jones N (2003). Challenging genome integrity. Biologija 1: 3–9.
Pašakinskienė I, Jones N (2005). A decade of ‘chromosome painting’ in Lolium and Festuca. Cytogenet Genome Res 109: 393–399.
Rapacz M, Gąsior D, Zwierzykowski Z, Leśniewska-Bocianowska A, Humphreys MW, Gay A (2004). Changes in cold tolerance and the mechanisms of acclimation of photosystem II to cold hardening generated by anther culture of Festuca pratensis × Lolium multiflorum cultivars. New Phytol 162: 105–114.
Scarth R, Law CN (1983). The location of the photoperiod gene, Ppd-B1 and additional genetic factor for ear emergence time on chromosome 2B of wheat. Heredity 51: 607–619.
Scarth R, Law CN (1984). The control of the day-length response in wheat by the group 2 chromosomes. Z Pflanzenzüchtg 92: 140–150.
Sim S, Chang T, Curley J, Warnke SE, Barker RE, Jung G (2005). Chromosomal rearrangements differentiating the ryegrass genome from the Triticeae, oat, and rice genomes using common heterologous RFLP probes. Theor Appl Genet 110: 1011–1019.
Sourdille P, Snape JW, Cadalen T, Charmet G, Nakata N, Bernard S et al (2000). Detection of QTLs for heading time and photoperiod response in wheat using a doubled-haploid population. Genome 43: 487–494.
Thomas HM, Harper JA, Meredith MR, Morgan WG, King IP (1997). Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome 40: 406–410.
Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E et al (1996). Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res 4: 486–490.
Thomas H, Morgan WG, Humphreys MW (1988). The use of a triploid for introgression in Lolium species. Theor Appl Genet 76: 299–304.
Thomas HM, Morgan WG, Humphreys MW (2003). Designing grasses with future – combining the attributes of Lolium and Festuca. Euphytica 133: 19–26.
Thomas HM, Morgan WG, Meredith MR, Humphreys WM, Thomas H, Leggett JM (1994). Identification of parental and recombined chromosomes in hybrid derivatives of Lolium multiflorum × Festuca pratensis by genomic in situ hybridization. Theor Appl Genet 88: 909–913.
Volkov RA, Panchuk II, Borysiuk LH, Borysiuk MB (2003). Plant rDNA: organisation, evolution, and using. Tsitol Genet 37: 72–78.
Yamada T, Jones ES, Cogan NOI, Vecchies AC, Nomura T, Hisano H et al (2004). QTL analysis of morphological, developmental, and winter hardiness-associated traits in perennial ryegrass. Crop Sci 44: 925–935.
Ziółkowski PA, Sadowski J (2002). FISH-mapping of rDNAs and Arabidopsis BACs on pachytene complements of selected Brassicas. Genome 45: 189–197.
Zwierzykowski Z, Lukaszewski A, Naganowska B, Leśniewska A (1999). The pattern of homoeologous recombination in triploid hybrids of Lolium multiflorum with Festuca pratensis. Genome 42: 720–726.
Zwierzykowski Z, Tayyar R, Brunell M, Lukaszewski AJ (1998). Genome recombination in intergeneric hybrids between tetraploid Festuca pratensis and Lolium multiflorum. J Hered 89: 324–328.
Zwierzykowski Z, Zwierzykowska E, Kosmala A, Łuczak M, Jokś W (2003). Genome recombination in early generations of Festuca pratensis × Lolium perenne hybrids. In: Zwierzykowski Z, Surma M, Kachlicki P (eds) Application of Novel Cytogenetic and Molecular Techniques in Genetics and Breeding of the Grasses. Institute of Plant Genetics PAS: Poznań. pp 63–69.
Acknowledgements
We thank Dr Hanna Weiss for the plasmid SK+ with 18S–25S rDNA and Dr Przemysław and Dr Katarzyna Nuc for the clone pU5SLL. We also thank MSc Włodzimierz Zwierzykowski and Magdalena Kapczyńska for technical support. This research was carried out in the frame of the European Union project entitled ‘Sustainable Grasslands withstanding Environmental Stresses’ (SAGES) under contract no. QLK5-CT-2000-00764.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kosmala, A., Zwierzykowski, Z., Gąsior, D. et al. GISH/FISH mapping of genes for freezing tolerance transferred from Festuca pratensis to Lolium multiflorum. Heredity 96, 243–251 (2006). https://doi.org/10.1038/sj.hdy.6800787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800787
Keywords
This article is cited by
-
Comparative analyses of DNA repeats and identification of a novel Fesreba centromeric element in fescues and ryegrasses
BMC Plant Biology (2020)
-
Cytogenetic and molecular genotyping in the allotetraploid Festuca pratensis × Lolium perenne hybrids
BMC Genomics (2019)
-
Karyotype reshufflings of Festuca pratensis × Lolium perenne hybrids
Protoplasma (2018)
-
Effects of introgressions from Festuca pratensis on winter hardiness of Lolium perenne
Euphytica (2017)
-
Similarities and differences in the nuclear genome organization within Pooideae species revealed by comparative genomic in situ hybridization (GISH)
Journal of Applied Genetics (2017)