Abstract
Hieracium pilosella (Asteraceae) was accidentally introduced to New Zealand about 100 years ago. Since then it has become an aggressive weed, and an unexpected degree of genetic and genome size variation has been detected; features that might result from interspecies hybridization. We investigated the possibility that H. pilosella has hybridized with related taxa. Of the four other subgenus Pilosella species introduced to New Zealand, H. praealtum is the most abundant and, on morphological and distributional evidence, most likely to be the other parent. Flow cytometry was used to estimate relative genome size for 156 Hieracium plants collected from the wild. Plants assigned to either parental or hybrid morphotypes were found to comprise tetraploid and pentaploid individuals using genome size measurements, and this was confirmed with direct mitotic chromosome counts for a subset of plants. The haploid DNA content of H. praealtum was approximately 22% larger than that of H. pilosella. Putative hybrids that were tetraploid had mean genome sizes equivalent to two H. pilosella and two H. praealtum haploid chromosome sets, implying they were hybrids arising from the fertilization of two reduced gametes. Similar results were obtained from tetraploid hybrids produced by controlled pollination. However, the majority of field hybrids were pentaploid with a genome size equivalent to four H. pilosella and one H. praealtum haploid chromosome sets. We infer that these are not first-generation hybrids but represent successful backcrossing with H. pilosella and/or hybrid–hybrid crossing, and that sexual tetraploid hybrids have been the parents. We note that populations putatively of H. pilosella often comprise apomictic pentaploid hybrids. Significantly, our data indicate the emergence of sexual hybrids that provide further opportunity for gene flow among taxa in this complex.
Similar content being viewed by others
Introduction
Gregor Mendel's pioneering genetic research was frustrated when he attempted to transfer his crossing experiments to Hieracium subgenus Pilosella. Mendelian genetics is difficult to demonstrate with this genus because it comprises species with extensive morphological and reproductive diversity that we now know consist of polyploid complexes maintained by facultative apomixis. For example, H. pilosella L. in Europe comprises plants with ploidies ranging from diploid to octoploid; the most common forms being tetraploid and pentaploid, and plants can be either obligate sexuals or facultative apomicts (Gadella, 1987, 1991; Krahulcova et al, 2000).
Since their accidental introduction into New Zealand about 100 years ago, H. pilosella and a number of other Hieracium species have become aggressive weeds in the high country of the South Island (over 200 m above sea level; Jenkins, 1991; Duncan et al, 1997). Members of the genus Hieracium have become the dominant taxa in over 500 000 ha of land (Hunter, 1991), with the mat-forming H. pilosella dominating large areas of endemic tussock grassland (Espie, 2001). Five stoloniferous species of Hieracium (subgenus Pilosella) are present in New Zealand and because they have similar or overlapping ecological requirements all can co-occur, but H. pilosella is the most abundant (Hunter, 1991). Of the five, H. pilosella is distinctive in having a single-stemmed flower stalk bearing a solitary capitulum with yellow ligules. In New Zealand, most individuals of H. pilosella are pentaploid; less commonly tetraploid and rarely hexaploid, heptaploid or octoploid occur. Plants with rare ploidies are found among pentaploid individuals (Jenkins and Jong, 1997; Chapman and Lambie, 2000). Tetraploid and pentaploid forms of H. praealtum Villars ex Gochnat have been found in New Zealand (Jenkins and Jong, 1997), often growing together with H. pilosella (Connor, 1992; Duncan et al, 1997). Both species reproduce predominantly via vegetative growth and gametophytic apomixis (of the aposporous type). However, most New Zealand H. pilosella plants are facultative apomictics, and seed production via (residual) sexual recombination can rise to 2.3% (Houliston and Chapman, 2001). Rare obligate sexual H. pilosella plants have been found in New Zealand, and these are predominantly tetraploid. Obligate sexual H. praealtum have not been recorded. Both species produce abundant viable pollen, although obligate sexual H. pilosella is self-incompatible (Gadella, 1991). Despite recent introduction and low levels of sexual reproduction, high levels of genetic variation have been detected using intersimple sequence repeat primers (ISSRs) in New Zealand populations of H. pilosella (Chapman et al, 2000). The hypothesis that part of this genetic diversity might have resulted from interspecific hybridization is the focus of the present study.
Although the gross karyotype of all Hieracium species is the same (n=9 chromosomes), a previous study using flow cytometry detected significant variation in haploid (1×) DNA content among several species of subgenus Pilosella (Bräutigam and Bräutigam, 1996; Bennett et al, 2000). Similar studies in other plant groups have shown DNA content to vary up to 200% without alterations of the basic karyotype (Brandham and Doherty, 1998), and this sort of variation in DNA content is difficult to assess or even identify using a microscope. Preliminary ploidy analysis of New Zealand populations of H. pilosella using flow cytometry detected some plants with DNA content intermediate in amount between that observed from tetraploid and pentaploid individuals. Such DNA content estimates might result from aneuploidy, indeed some aneuploid H. pilosella have previously been reported in New Zealand (Chapman and Lambie, 2000). However, the condition is regarded as uncommon in natural European populations (Bräutigam and Bräutigam, 1996), and was not detected in a cytogenetic study of 231 H. pilosella samples from New Zealand (Jenkins and Jong, 1997). An alternative explanation for the unusual genome size results is interspecific hybridization. Breeding experiments have shown that a variety of combinations of interspecific hybridization between H. pilosella, H. praealtum, H. aurantiacum L. and H. caespitosum Dumort. is possible in the glasshouse (Mendel, 1869; Chapman and Bicknell, 2000). Putative interspecific hybrids are observed in the wild in Europe (Krahulcova et al, 2001), and a number of hybrid taxa have been named (see for example Sell and West, 1976). Excluding the distinctive taxon H. × stoloniflorum which was almost certainly introduced as a hybrid (unpublished data), novel hybrids have not previously been reported in New Zealand. Based on flower colour and occurrence in New Zealand, the only species that might be the other parents of yellow flowered hybrids are H. praealtum, which is the most common and widespread, and H. caespitosum, which is much rarer (Webb et al, 1988; Duncan et al, 1997).
Flow cytometry has successfully been applied to the study of hybridization in a number of plant taxa including allotetraploid Glycine (Hammatt et al, 1991) and willow (Thibault, 1998), and homoploid hybrids of Limonium (Morgan et al, 1995). In Hieracium, the hybrid origin of two species (H. brachiatum and H. leptophyton) has been inferred from intermediate DNA content measures (Bräutigam and Bräutigam, 1996). Therefore, we undertook a study of genome size of H. pilosella, H. praealtum and putative hybrids from New Zealand using flow cytometry, complemented by direct chromosome counts of a subset of plants.
Materials and methods
Rationale
We propose that Hieracium plants with apparent interploid DNA content are interspecific hybrids, and that backcrossing with parental-taxa could explain the unexpectedly high degree of observed genetic variation within putative H. pilosella. Pentaploids can produce 5 ×, 3 × and 2 × pollen and ovules, and rare aneuploid gametes (Krahulcova and Krahulec, 2000), while tetraploid plants produce 2 × and rarely 4 × gametes (Gadella, 1988). Therefore, hybridization arising from the fertilization of two reduced gametes (BII hybridization) between H. pilosella and H. praealtum could theoretically result in predominantly tetraploid and pentaploid offspring. Subsequent progeny from outcrossing BII hybrids could potentially result in plants with a range of permutations of the two parental genomes. Given that pentaploid H. praealtum is rare in New Zealand, and the most likely maternal parents of hybrids are obligate sexual, tetraploid H. pilosella, F1 hybrids are expected to be tetraploid (Figure 1). A tetraploid hybrid, could theoretically contain either two chromosome sets from each parental taxon (2pil,2pra, where pil=H. pilosella and pra=H. praealtum) or one plus three chromosome sets from the parental taxa (1pil,3pra; 3pil,1pra). A pentaploid hybrid could be one of four possible combinations of parental haploid genomes: 4pil,1pra; 3pil,2pra; 2pil,3pra; 1pil,4pra (Figure 1). Hybridization involving the fusion of one reduced and one unreduced gamete (BIII hybridization) may result in 6 × and 7 × offspring, and this process is probably integral to the evolution of polyploid complexes in the genus. Indeed, BIII hybrids are fairly common in Europe, and many named taxa probably originate in this way.
The difference in size of the putative parental genomes is sufficient for us to distinguish between the genome sizes predicted from the seven likely gamete combinations using flow cytometry, and thus enables us to compare predicted values with those obtained from putative hybrid plants. In this way, we can obtain evidence for the hybridization of H. pilosella with other Hieracium species, the ploidy of the hybrids and their frequency.
Morphological identification
Plants that had a single capitulum per inflorescence and lower surfaces of their leaves pale grey or white due to the presence of numerous stellate hairs were identified as H. pilosella. Plants were coded as H. praealtum if they had multiple capitula per inflorescence, their capitula were smaller, leaves were narrower, more blue in colour and lacked stellate hairs on the underside. H. caespitosum, which is present but rare at a few of the sites we studied, was distinguished by the presence and distribution of many long simple hairs on the underside of leaves, and stellate hairs on the upper leaf surface, and these plants were excluded (Webb et al, 1988). Plants that had two or more capitula per inflorescence and stellate hairs on the lower surface of their leaves, were coded as ‘hybrid’.
Plant collection and locations
We examined 156 plants collected from 19 locations in New Zealand (Figure 2). This set comprised 53 H. pilosella, 67 hybrids (referred to as field hybrids) and 36 H. praealtum (Table 1). Most of the plants we identified as interspecies field hybrids came from two intensively studied locations, Drac Flat and Lyndon. These two populations have been used to investigate genetic structure and the origin of tetraploid plants in New Zealand using molecular markers (Chapman et al, 2000; Chapman et al, 2003). All plants were potted up in the glasshouse and grown under the same conditions of light, temperature and nutrition, where flowering and leaf morphology was noted.
Glasshouse cross pollination
Pollen from a single tetraploid H. praealtum plant was used to fertilize flowers from a tetraploid H. pilosella (DF1.1) and a tetraploid putative F1 hybrid (DF1.10). In each case, two capitula on two ramets of the maternal plants were pollinated at full flower opening and without emasculation. Both maternal ramets originated from Drac Flat and were cultivated in the glasshouse. The seeds produced were cultivated, and a random sample of the resultant seedlings were grown on for further study.
Flow cytometry
Isolation of nuclei from leaf tissue followed the method of Galbraith et al (1983) with some modifications. A fragment of fresh leaf tissue (∼6 mm2) was placed together with a similar amount of leaf from the reference species used as an internal standard, in a plastic Petri dish. Leaf tissue was chopped in the presence of one drop of commercial nuclei isolation buffer, UV CyStain Precise T solution A (100 ml deionized water, 2.1 g citric acid, 0.5 g Tween 20) (PARTEC GmbH, Münster, Federal Republic of Germany) using a fresh stainless-steel razor blade. When well chopped an additional 0.5 ml of solution A was added. After approximately 3 min the sample was filtered through a 30-μm filter, and 2.0 ml of Partec Cystain Precise T solution B (100 ml deionised water, 7.9 g dibasic sodium phosphate, 0.5 ml DAPI stock (4.55 mg 4′, 6′-diamidino-2-phenylindole, 10 ml deionised water) was added. Samples were analysed for DNA content after at least 3 min of staining. For this, the Partec PA-II Particle Analysing System (PAS) was employed, using filter combinations of UG 1, TK420, TK590, and GG435 and a mercury arc lamp (HBO 100 W/2). The standard used was the diploid Asteraceae species Bellis perennis L., and we calculated a calibration factor using a tetraploid H. pilosella in order to estimate ploidy of subsequent H. pilosella samples from the index obtained during each run. For each sample 5000 nuclei were measured. In most instances, each plant was screened twice on separate occasions. DNA mass estimates were only accepted when their coefficient of variation (CV) was <4%. The average CV for the 156 plants was 2.59%. DNA was stained with DAPI, a nonintercalating dye that binds preferentially to A–T bases and results cannot therefore be directly expressed in picograms (Johnston et al, 1999). We subsequently also obtained representative measurements of DNA content using a clone of the diploid H. peleterianum as an internal standard instead of Bellis perennis. Relative measures of genome size using a diploid member of the genus may be more practical for comparisons with future studies of the group.
Ploidy analysis
Initially, we followed our procedure for inferring the ploidy of H. pilosella from DNA content measurement. We used the conversion factor based on the flow peak ratio obtained from multiple sampling of a known tetraploid H. pilosella with an internal standard (see above), to estimate chromosome number (and thus ploidy) of subsequent plants by dividing each peak ratio value obtained by the conversion factor. For individuals of H. pilosella this resulted in values of approximately 36 (ie 4 ×) or 45 (ie 5 ×). Inferred chromosome numbers for other species (eg H. praealtum) appeared higher as a result of the overall larger genome size, and as noted above, some morphologically intermediate plants gave apparently intermediate inferred chromosome numbers (typically about 40). These H. pilosella-relative chromosome scores were used to plot size frequency graphs (eg Figure 3), and we used the mean scores obtained from morphologically stable tetraploid H. pilosella and H. praealtum plants to calculate expected values for seven possible hybrid genome permutations (Figure 1). We assumed that there is no recombination between the chromosomes derived from the two parental species.
In addition, for a subset of plants from the three morphological categories plus individuals of other species, ploidy was determined using chromosome counts. Mitotic chromosome counts of squashed root tip preparations used lacto-propionic orcein staining following the method of Krahulcova and Krahulec (1999). For plants analysed in this manner, we used the peak ratios obtained from flow cytometry to calculate a relative 1 × DNA content for comparison among taxa and ploidy levels (Table 2).
Results
Flow cytometry
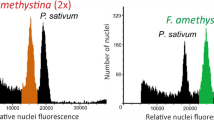
The size of the genomes of H. pilosella, H. praealtum and putative hybrids examined varied considerably. In H. praealtum the inferred haploid (1×) genome was slightly larger in tetraploids than in pentaploids (see Table 2), and was approximately 22% larger than the genome of H. pilosella. The frequency chart (Figure 3) reveals two discrete size ranges for DNA measurements of each taxon, corresponding to tetraploids and pentaploids. The smallest nuclei size class comprised 33 tetraploid H. pilosella. Tetraploid hybrid plants (n=16) form the second discrete size class, and tetraploid H. praealtum (n=29) the third. The majority of field hybrids (n=50) had DNA measurements with a range that partially overlaps pentaploid H. pilosella (n=20). Although plants in these two groups have similar genome sizes (Figures 3 and 4), their means differ significantly (P<0.001). The largest nuclei are those of pentaploid H. praealtum plants (n=8).
Boxplots showing mean and range (75% and ∼95%) of genome sizes, expressed as H. pilosella-relative chromosome number, for H. pilosella, H. praealtum and field hybrids. The predicted sizes of seven theoretical hybrid genome permutations are indicated for comparison. Size distributions of hybrids are indicated by black fill.
Of the seven theoretical hybrid haploid-genome combinations, five have predicted means that fall outside of the genome size ranges detected (3pil,1pra; 1pil,3pra; 3pil,2pra; 2pil,3pra; 1pil,4pra: see Figure 4). Our data indicate that the tetraploid hybrids consist of two H. pilosella haploid genomes and two H. praealtum haploid genomes (2pil,2pra) with a predicted mean of 40.65, compared to the observed mean of 40.71. The pentaploid hybrids most probably have the 4pil,1pra combination of chromosomes with a predicted mean genome size of 47.83 and an observed mean of 47.34 (Figures 3 and 4).
Chromosomes
Chromosome counts of the two likely parental species (H. pilosella, H. praealtum) revealed tetraploid and pentaploid plants only, and no aneuploids (Table 2). Among the field hybrid plants there were also tetraploids and pentaploids, indicating they were all derived from reduced gametes (BII hybrids) rather than from the fusion of unreduced and reduced gametes (BIII hybrids) as seen by Bräutigam and Bräutigam (1996). In all cases, chromosome counts agreed with ploidy determined by DNA measurements using flow cytometry, and corroborated our inferences.
All H. praealtum and the single H. caespitosum examined contained in their complement one chromosome that appeared larger than any of the others, as has been previously reported for these and some related taxa but not for H. pilosella (Jenkins and Jong, 1997; Krahulcová and Krahulec, 1999). We did not find this large ‘marker’ chromosome in any individuals of H. pilosella or the pentaploid hybrids, but it was present in three of the 12 tetraploid hybrid plants examined (Table 2). Three of the tetraploid hybrids from Lyndon had unusual karyotypes with additional ‘fragments’ or broken chromosomes (Table 2).
Glasshouse hybrids
In all, 27 seedlings cultivated from controlled cross-pollination were subjected to flow cytometry. These individuals had DNA contents intermediate between H. pilosella and H. praealtum, and similar to scores obtained from tetraploid field hybrids collected. The nine seedlings from DF 1.1× H. praealtum had a narrower size range than the 18 seedlings from DF 1.10× H. praealtum. This probably results from different permutations of ancestral genomes in gametes from DF 1.10 (the putative F1 hybrid). In particular, two individuals at either end of the range had DNA contents consistent with the genome permutations 3pil,1pra and 3pra,1pil (Figure 5).
Frequency chart of genome size estimates, expressed as H. pilosella-relative chromosome number, from flow cytometry of (a) hybrid Hieracium from the field, and (b) the progeny of two glasshouse crosses between H. pilosella 4× mother × H. praealtum 4× father, and (c) field hybrid 4× mother × H. praealtum father. Arrows on (c) indicate DNA content estimates that correspond to size classes expected from (×) 3pil,1pra and (y) 1pil,3pra genomes.
Discussion
DNA content of Hieracium taxa
While the karyotypes of H. pilosella and H. praealtum are not sufficiently different in size to be distinguishable using standard microscopic studies of mitotic cells, using flow cytometry, we have confirmed that H. praealtum haploid genomes have approximately 22% more DNA than H. pilosella. In addition, a single large marker chromosome is diagnostic for H. praealtum and H. caespitosum (see Jenkins and Jong 1997; Krahulcova and Krahulec, 1999).
Our single estimate of 1× DNA content from a tetraploid H. caespitosum was the same as the mean 1× score from pentaploid H. praealtum. Our H. × stoloniflorum and H. aurantiacum had DNA contents higher than H. pilosella but lower than H. caespitosum and H. praealtum. This is consistent with the observations of Bräutigam and Bräutigam (1996) with respect to H. aurantiacum, but the relative haploid DNA content of their tetraploid H. × stoloniflorum was larger than our hexaploid H. × stoloniflorum which reflects the multiple origins of this hybrid species. The inferred 1× DNA content of tetraploid and pentaploid H. pilosella differed little, while in H. praealtum the 1× DNA content of tetraploids was substantially larger (∼3.5%) than pentaploids in our sample, which we have no explanation for. Similarly, the 1× DNA content of tetraploid field hybrids was substantially larger (∼7%) than pentaploid hybrids, probably because of their composite nature.
Identification of hybrids
In the field in New Zealand, putative hybrids are not readily distinguished from H. pilosella at the same locations. But, after growing in glasshouse conditions, leaf morphology and capitula number differentiate and allow their identification. While flow cytometry can be used to distinguish the two common ploidy levels (tetraploid and pentaploid) of H. pilosella and H. praealtum, overlap between pentaploid H. pilosella and pentaploid field hybrids could result in incorrect identification using this method alone. However, the clear difference in genome size of tetraploid plants has allowed us to infer that hybridization has occurred in the field between H. pilosella and (most probably) H. praealtum. Without the use of taxon-specific molecular markers it is not possible to exclude other related species, but rarity and difference in DNA content (eg 4× H. caespitosum, see Table 2) indicate that it is unlikely that other species were involved. For the same reasons, it is unlikely that pentaploid H. praealtum have contributed to the hybrids examined. We found no evidence of BIII (addition) hybrids as observed in Europe by Bräutigam and Bräutigam (1996), where unreduced pollen from pentaploid H. praealtum yield hybrids.
In the present study, the 12 tetraploid field hybrids have genome sizes indicative of two haploid genomes from each parental species. These plants are probably F1 hybrids, but could be derived from a subsequent generation if there is no recombination among the parental chromosome sets. The pentaploid hybrids have a mean DNA content consistent with them comprising four H. pilosella haploid chromosome sets plus one H. praealtum haploid set. It is unlikely that this combination could result from a first-generation hybridization event and thus we infer that backcrossing and/or interhybrid-crossing has occurred. Only two of the seven possible hybrid genome combinations were encountered, suggesting other combinations are less successful in the field.
In the glasshouse, hybrids generated from known parents by controlled pollination have the expected intermediate DNA content equivalent to the 2pil,2pra theoretical composite genome. We note that the progeny from our cross of tetraploid H. praealtum with a tetraploid field hybrid (2pil,2pra) as the mother, comprise a wider range of DNA contents than the progeny of a tetraploid H. pilosella (4pil) mother. This is to be expected as the 2pil,2pra genome can generate gametes with a number of different genome permutations. Further replicates are needed to confirm these observations.
Sexuality, invasion and incipient speciation
We noted a wide range of morphotypes among the tetraploid field hybrids examined with considerable variation in leaf shape and colour, density and combination of hairs, number and size of flowers, and proportion of apomictic seed set. This may be a result of different parental individuals being involved, or indicate the polymorphic nature of the parent genome, such that gametes carry different alleles. The latter is likely as the parent taxa are polyploids and themselves the products of (more ancient) interspecies or interlineage hybridization.
Recombination could also explain this morphological variation but our DNA measurements show that genome size of plants collected from the field form discrete size classes suggesting that recombination is not common between the genomes of these species. Either hybridization has happened recently in New Zealand from pure parental stock or recombination between parent-taxa genomes within hybrid plants is infrequent.
One biologically important implication of the existence of 4pil,1pra pentaploid hybrids is that at least some F1 hybrids are conduits for gene flow. Because pentaploid H. pilosella are facultative apomictic, but produce both 2× and 3× pollen, it is probable that F1 hybrids have acted as sexual, maternal parents in backcrossing. Preliminary observations of field hybrids maintained in the glasshouse indicate that the degree of apomixis among tetraploid (F1) hybrids varies considerably. Some individuals appear to produce ∼50% apomictic seed (ie without fertilization), while others produce none. While it is possible that unfilled seed is simply an indication of sterility, preliminary glasshouse experiments using pentaploid H. pilosella or tetraploid H. praealtum as a pollen donor to field hybrids (that set no or very few apomictic seed) results in the formation of full viable seed, implying that these tetraploid hybrids are obligate sexuals. The recovery of sexual hybrids may have a substantial effect on continuing hybridization given the relative scarcity of obligate sexual (normally tetraploid) H. pilosella in New Zealand.
This sexuality appears to have facilitated the formation of H. pilosella-like pentaploids with the potential benefits of additional alleles inherited from H. praealtum. As these pentaploid hybrids are facultative apomicts they have the power to disperse and easily colonize new habitats. We note that many smaller, adventive populations of putative-H. pilosella, actually consist of apomictic pentaploid hybrids with intermediate morphology, and larger populations also comprise a component of these forms. Such populations probably comprise one or few clones derived from seed colonization and their existence could mark the emergence of new lineages of Hieracium in New Zealand subject to selection in a range of habitats. The success of H. pilosella (and its hybrids) in New Zealand as an invasive weed may owe much to the genetic variability it has been able to incorporate via intra- and interspecific crossing. Work is underway to develop molecular markers to test the hybridization hypothesis and investigate the extent of introgression.
The generation of hybrids in New Zealand is of interest to evolutionary and conservation biologists. Hybridization is likely to result in the generation of novel genotypes with high additive genetic variance (Lee, 2002). This is likely to increase the possibility of invasion in a greater range of native habitats, and may reduce the effectiveness of biological control measures (Ellstrand and Schierenbeck, 2000). However, the phenomenon also promises to illuminate our knowledge of plant speciation processes. At the very least this will assist in understanding the complex history of native Hieracium in Europe, but promises to provide a fruitful model system for the study of the role of polyploidy and hybridization in species formation (Abbott, 1992; Soltis and Soltis, 2000). Hieracium is of particular interest because there is combined within single taxa (or polyploid species complexes) a range of reproductive strategies (probably controlled by a single locus; Chapman and Bicknell, 2000) that ought to provide ample opportunity for the capture and fixation of genetic diversity.
As Mendel observed in a letter to Carl Nägeli, ‘the naturally occurring hybridization in Hieracium ……. would finally result in the disappearance of the species involved, while one or another of the more happily organised progeny, better adapted to the prevailing telluric and cosmic conditions, might take up the struggle for existence successfully’ (Orel, 1996).
References
Abbott RJ (1992). Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol 7: 401–405.
Bennett MD, Bhandol P, Leitch IJ (2000). Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Ann Bot 86: 859–909.
Brandham PE, Doherty M-J (1998). Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot 82 (Suppl A): 67–73.
Bräutigam S, Bräutigam E (1996). Determination of the ploidy level in the genus Hieracium Subgenus Pilosella (Hill) S.F. Gray by flow cytometric DNA analysis. Folia Geobot Phyotax 31: 315–321.
Chapman HM, Bicknell R (2000). Recovery of a sexual and an apomictic hybrid from crosses between facultative apomicts Hieracium caespitosum and H. praealtum. NZ J Ecol 24: 81–85.
Chapman HM, Lambie SC (2000). Chromosome numbers in New Zealand populations of Pilosella officinarum F.W. Shultz. & Sch. Bip. IOPB Newslett 31: 12.
Chapman HM, Robson B, Ilene I (2003). A case of reversal: the evolution and maintenance of sexuals from parthenogenetic clones in Hieracium pilosella. Int J Plant Sci 164: 719–728.
Chapman HM, Parh D, Oraguzie N (2000). Genetic structure and colonizing success of a clonal, weedy species, Pilosella officinarum (Asteraceae). Heredity 84: 401–409.
Connor HE (1992). Hawkweeds, Hieracium spp. in tussock grasslands of Canterbury, New Zealand, in 1960s. NZ J Bot 30: 247–261.
Duncan RP, Colhoun KM, Foran BD (1997). The distribution and abundance of Hieracium species (Hawkweeds) in the dry grasslands of Canterbury and Otago. NZ J Ecol 21: 51–62.
Ellstrand NC, Schierenbeck KA (2000). Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA 97: 7043–7050.
Espie P (2001). Hieracium in New Zealand: Ecology and Management. AgResearch Ltd: Lincoln, New Zealand.
Gadella TWJ (1987). Sexual tetraploid and apomictic pentaploid populations of Hieracium pilosella (Compositae). Plant Syst Evol 157: 219–245.
Gadella TWJ (1988). Some notes on the origin of polyploidy in Hieracium pilosella aggr. Acta Bot Neerl 37: 515–522.
Gadella TWJ (1991). Variation, hybridization and reproductive biology of Hieracium pilosella L. Proc Kon Ned Akad v Wetensch 94: 455–488.
Galbraith DW, Harkins KR, Maddoz JM, Ayres NM, Sharma DP, Firoozabady E (1983). Rapid flow cytophotometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051.
Hammatt N, Blackhall NW, Davey MR (1991). Variation in the DNA content of Glycine species. J Exp Bot 42: 659–665.
Houliston GJ, Chapman HM (2001). Sexual reproduction in field populations of the facultative apomict, Hieracium pilosella. NZ J Bot 39: 141–146.
Hunter G (1991). The distribution of Hawkweeds (Hieracium) in the South Island, indicating a problem status. J NZ Mountain Lands Inst 48: 1–10.
Jenkins TA (1991). A review of characteristics of mouse-ear hawkweed (Hieracium pilosella). NZ J Ecol Soc Occ Pub No 2: 15–23.
Jenkins TA, Jong K (1997). Significance of polyploid variation in New Zealand Pilosella and Hieracium (Asteraceae). Bot J Scotl 49: 75–87.
Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ (1999). Reference standards for determination of DNA content of plant nuclei. Am J Bot 86: 609–613.
Krahulcova A, Krahulec F (1999). Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in the Krkonose Mts (the Sudeten Mts). Preslia 71: 217–234.
Krahulcova A, Krahulec F (2000). Offspring diversity in Hieracium subgen. Pilosella (Asteraceae): new cytotypes from hybridization experiments and from open pollination. Ftagm Flor Geobot 45: 239–255.
Krahulcova A, Krahulec F, Chapman HM (2000). Variation in Hieracium subgen. Pilosella (Asteraceae): what do we know about its sources? Folia Geobot 35: 319–338.
Krahulcova A, Krahulec F, Chrtek JJ (2001). Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in Krkonose Mts (the Sudeten Mts)-2. Preslia 73: 193–211.
Lee CE (2002). Evolutionary genetics of invasive species. Trends Ecol Evol 17: 386–391.
Mendel G (1869). Ueber einige aus künstliche Befruchtung entnommenen Hieracium - Bastarde. Verh naturf Ver Brünn 8: 26–31.
Morgan ER, Burge GK, Seelye JF, Grant JE, Hopping ME (1995). Interspecific hybridization between Limonium pergrinum Bergius and Limonium purpuratum L. Euphytica 83: 215–244.
Orel V (1996). Gregor Mendel: the First Geneticist. Oxford University Press: Oxford.
Sell PD, West C (1976). Hieracium L. In: Tutin TG, Hewood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA (eds) Flora Europeae, Vol 4, Plantaginaceae to Compositae (and Rubiacieae). Cambridge University Press: Cambridge, pp 358–410.
Soltis PS, Soltis DE (2000). The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci USA 97: 7051–7057.
Thibault J (1998). Nuclear DNA amount in pure species and hybrid willows (Salix): a flow cytometric investigation. Can J Bot 76: 157–165.
Webb CJ, Skyes WR, Garnock-Jones PJ (1988). Flora of New Zealand. Vol IV. DSIR Botany Division: Christchurch, New Zealand.
Acknowledgements
This work was supported by grants from Canterbury University (UOC Research Grants U6435 & U2022). We thank Ross Bicknell, Gary Houliston and Beth Robson, for their help in the laboratory, glasshouse and field. Scott Dunavan provided additional plants. Comments from two reviewers improved this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morgan-Richards, M., Trewick, S., Chapman, H. et al. Interspecific hybridization among Hieracium species in New Zealand: evidence from flow cytometry. Heredity 93, 34–42 (2004). https://doi.org/10.1038/sj.hdy.6800476
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800476
Keywords
This article is cited by
-
Apomixis and genetic background affect distinct traits in Hieracium pilosella L. grown under competition
BMC Biology (2021)
-
Gymnocalycium ×applanatum (Cactaceae, Cactoideae) – the first reported nothospecies between the subgenera Gymnocalycium and Trichomosemineum
Folia Geobotanica (2021)
-
Invasion by hawkweeds
Biological Invasions (2021)
-
Dynamics of apomictic and sexual reproduction during primary succession on a glacier forefield in the Swiss Alps
Scientific Reports (2020)
-
Unusual suspects in the usual places: a phylo-climatic framework to identify potential future invasive species
Biological Invasions (2017)