Abstract
The reconstruction of recent historical population sizes allowed us to investigate the influence of random evolutionary processes on present-day genetic diversity in populations of Dryopteris cristata. This long-lived, allotetraploid fern is rare and endangered in the study area at the southwestern border of its European distribution. Random amplified polymorphic DNA (RAPD) diversity of 280 individuals from 14 populations of D. cristata was extraordinarily low, suggesting an ancient bottleneck in the species’ history. Analysis of molecular variance (AMOVA) of 25 different RAPD multiband phenotypes revealed significant genetic variation among three geographical regions (15%) and among populations within regions (34%); 51% of total variance was attributed to variation within populations. High population differentiation indicated limited gene flow among populations, and genetic divergence was not correlated with geographical distance. There was no relationship between genetic variation within population, estimated as molecular variance, and present-day population size. Populations with recent historical bottlenecks of fewer than 25 individuals showed a substantial and significant reduction in genetic variation, compared with populations without bottlenecks. Comparatively high levels of genetic variation were still maintained in small remnants (60–110 individuals) of formerly large populations. Average deviations of frequencies of widespread polymorphic markers within populations from their frequencies in the whole dataset were significantly higher in small or recently bottlenecked populations than in constantly large populations, thus providing evidence for random sampling effects during genetic bottlenecks and drift in small populations. The present investigation demonstrates the importance of population history for understanding present-day genetic diversity within natural populations, as well as for conservation biology.
Similar content being viewed by others
Introduction
The genetic diversity in a species or population is the result of its evolutionary history and of recent evolutionary processes. Low levels of genetic diversity may reduce the potential of species or populations to survive in a changing environment (Ellstrand & Elam, 1993; Lande & Shannon, 1996). Theory predicts that genetic drift, increased inbreeding and reduced gene flow will all decrease genetic diversity in small and isolated populations. Hence, a paradigm in conservation biology states that genetic diversity decreases with decreasing population size (Barrett & Kohn, 1991; Young et al., 1996). A positive correlation between actual population size and genetic diversity has been found in several plant species (Godt et al., 1996; Fischer & Matthies, 1998). In the locally rare Salvia pratensis and Scabiosa columbaria, population size was correlated with the proportion of polymorphic allozyme loci and the mean number of alleles, but no correlation was detected between population size and gene diversity (van Treuren et al., 1991). No relationship was reported between population size and genetic diversity for other species (Ellstrand & Elam, 1993; Lutz et al., 2000; Schmidt & Jensen, 2000). Historical changes in population size and isolation were proposed as possible explanations for the lack of correlation in the latter studies. However, because information on recent population history is scarce, it is usually not possible to test its influence on genetic diversity (Lutz et al., 2000; Schmidt & Jensen, 2000).
In the short term, loss of genetic diversity in long-lived plants is supposedly the result of severe genetic bottlenecks rather than genetic drift in small populations (Young et al., 1996). Both processes were summarized as ‘random sampling effects’ by Barrett & Kohn (1991). Random sampling is a single event during a genetic bottleneck, but it accumulates during genetic drift in any extended period of small population size. Barrett & Kohn (1991) stated that theoretical work on stochastic loss of genetic diversity is based largely on a diploid, outbreeding system, which might be unrepresentative of many plant species. A theoretical prediction of genetic drift is that allele frequencies within populations change erratically and tend to drift apart, eventually resulting in the fixation and loss of alleles, while overall average allele frequencies among populations remain constant (Hartl & Clark, 1997). Increased genetic differentiation is expected among populations affected by random sampling. Furthermore, theory states that a genetic bottleneck primarily eliminates low frequency alleles, while the remaining alleles tend to persist in intermediate frequencies within the population (Nei et al., 1975). Genetic bottlenecks can be caused by a sharp reduction in the number of individuals (population bottleneck) or by the establishment of a new population by a few founder individuals (founder effect). Recent genetic bottlenecks have been demonstrated in introduced populations of Sarracenia purpurea (a single founder individual; Schwaegerle & Schaal, 1979) and Argyroxiphium sandwicense ssp. sandwicense (reintroduction with seeds from two individuals; Robichaux et al., 1997; Friar et al., 2000).
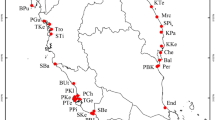
Switzerland has a long tradition of floristic research. The reconstruction of the former distribution of Dryopteris cristata, a locally rare fern in Switzerland, and, for most of the remaining populations, the reconstruction of changes in population size during the past 120 years, was thus possible from literature and herbaria (Landergott et al., 2000). In addition, the present and former distribution of D. cristata has been studied in the neighbouring area of Baden-Württemberg in southern Germany (Willbold, 1995). In both areas, D. cristata inhabits alder woodland and formerly exploited peat bogs (Willbold, 1995; Landergott et al., 2000). Such habitats are naturally isolated and mostly of restricted size, but have been further fragmented and isolated as a consequence of habitat destruction prior to 1945 (Fig. 1; Landergott et al., 2000). The economic exploitation of peat bogs often caused distinct changes in population sizes of D. cristata. Three trends in recent population history could be distinguished: (i) the reduction of a large population to a small remnant before 1945; (ii) the increase of a formerly small population to a presently large one; (iii) a severe historical bottleneck of fewer than 25 individuals (Table 1; Landergott et al., 2000). This categorization allowed investigation into how random sampling in recent population history has affected present-day genetic diversity in natural populations of D. cristata. Additionally, the evaluation of genetic diversity and population differentiation is of importance for this species’ conservation.
Present and former distribution of populations of Dryopteris cristata in Baden-Württemberg in southern Germany, Liechtenstein and Switzerland, according to Willbold (1995) and Landergott et al. (2000), with locations of the 14 populations sampled for RAPD analysis (for population abbreviations, see Table 1).
Random amplified polymorphic DNA (RAPD) fragments (Weising et al., 1995) are considered to be selectively neutral markers (Bachmann, 1997) and are thus useful for studying genetic differentiation and the effects of random sampling. In the present study, the following questions were addressed by investigating 14 populations of D. cristata using RAPD markers:
1 Is there significant genetic differentiation among populations or among regions, and is genetic divergence correlated with geographical distance?
2 Are large populations genetically more variable than small ones?
3 Is the present-day genetic diversity affected by recent population history? In particular, have historical bottlenecks of fewer than 25 individuals decreased the genetic diversity in natural populations of D. cristata?
Materials and methods
Study species
Dryopteris cristata (L.) A. Gray is a long-lived, lowland, homosporous fern of wet bogs and swampy woodland with a temperate to subarctic, circumpolar distribution (Dostal et al., 1984); the Alps are the southern border of its European distribution. The species shows little morphological variation throughout its range. In a comparison of North American and European plants (Walker, 1961), all individuals were cytologically uniform and resembled each other morphologically and ecologically. Furthermore, an allozyme investigation of eight German populations of D. cristata detected no genetic diversity (Güldenpenning, 1994), while an allozyme survey of 15 populations from the British Isles, Austria and Switzerland revealed only little polymorphism, with most genetic diversity among populations (J. C. Vogel, Natural History Museum, London, unpubl. data).
Dryopteris cristata has become a rare species in southwestern Central Europe and is considered to be endangered according to the Red Data Books of Austria, France, Germany and Switzerland (Landergott et al., 2000).
Dryopteris cristata, D. carthusiana (Vill.) Fuchs and D. dilatata (Hoffm.) Gray (Aspidiaceae) form a complex of three closely related allotetraploids (2n=164), sharing ancestral diploid genomes (Walker, 1961). One parent of D. cristata is the diploid North American species, D. ludoviciana (Kunze) Small (Walker, 1969). The other parent of D. cristata, which it shares with D. carthusiana, is a hypothetical ancestral diploid referred to D. semicristata (Werth, 1989).
Sampling
Fourteen populations of D. cristata in Switzerland, Liechtenstein and Baden-Württemberg in southern Germany were investigated (Fig. 1). According to present-day gaps in the distribution of D. cristata, the study area was divided into three geographical regions (Table 1): western Switzerland (W), eastern Switzerland (E) and Baden-Württemberg (B). Six large populations with more than 300 individuals and six small ones with fewer than 150 individuals were chosen among these three regions. All the remaining populations in western Switzerland (W1–W4; Fig. 1) — three small ones and a large one — were sampled. Two additional populations from eastern Switzerland (*E5 and *E6), most probably founded in the second half of the 20th century (Landergott et al., 2000), were sampled as well.
In each of the 14 populations, fresh leaf material of 20 individuals was randomly sampled throughout the population area in July 1999. Leaf material was immediately dried in silica gel and subsequently stored at −80°C. Population size was counted for up to 300 individuals, beyond this it was estimated into two classes: 300–500 (<500) and more than 500 individuals (>500; Table 1).
RAPD analysis
Genomic DNA was extracted using the FastDNA Kit (Bio 101) following the manufacturer’s protocol. In an additional purification step, DNA was precipitated with 1/10 volume of 3 M sodium-acetate solution (pH 5.2) and two volumes of cold ethanol (96%), precipitated again with two volumes of cold ethanol (70%) and finally stored in TE buffer. After optimization of reaction mixtures and conditions for thermal cycling, PCR amplifications were carried out in 12.5 μL reaction volumes of 2 mM MgCl2, 0.1 mM of each dNTP, 1× the supplied polymerase buffer, 0.04 U μL–1 Taq DNA polymerase (Amersham Pharmacia Biotech), 0.2 μM single or 0.3 μM pairwise oligonucleotide RAPD-primers (Operon Technologies) and approximately 3 ng of template DNA. The following cycles were run on a Genius thermal cycler (Techne): initial denaturation for 3 min at 94°C, 40 cycles of 30 s/94°C, 60 s/39°C, 90 s/72°C and a final extension of 5 min/72°C. Amplification products were separated on 1.5% agarose gels in 1× TAE buffer at 65 V for 4.5 h, stained with ethidium bromide and photographed under UV light.
Primer screening was performed using 80 primers (Operon Technologies kits A–D) and DNA samples of 12 individuals, one from each population (except *E5 and *E6). Seventy-three of these primers yielded amplification products. In addition, 66 pairwise combinations of primers, which gave uniform patterns when used as single primers, were screened in two individuals. Essentially new banding patterns were obtained with eight of these primer pairs. Hence, 10 individuals, each from a different population, were screened for polymorphic markers with these eight primer combinations as well.
Negative controls were run in order to confirm that no artefacts or contamination occurred. Being aware of the method’s susceptibility to changes in reaction conditions (Skroch & Nienhuis, 1995; Weising et al., 1995), reproducibility of all variable fragments was confirmed by several repeated amplification reactions, and by additional amplifications of two to six samples using slightly varying reaction mixtures, i.e. varying concentrations of template DNA (1–6 ng per reaction volume) and MgCl2 (1.5–3 mM). Finally, the whole procedure including DNA extraction was repeated for six individuals. As a result of these tests, one polymorphic fragment produced by primer OPC-20 had to be excluded because it was not reproducible under varying MgCl2 concentrations.
Statistical analysis
Presence or absence of RAPD fragments was scored visually and recorded in a binary data matrix. RAPDs are dominant markers and therefore analysis of multiband phenotypes has been recommended rather than analysis of estimated allele frequencies, which requires basic assumptions including Hardy–Weinberg equilibrium (Bachmann, 1997). Variation in RAPD data was analysed with hierarchical analysis of molecular variance (AMOVA; Excoffier et al., 1992), based on pairwise squared Euclidean distances among RAPD phenotypes. Input files for the AMOVA V.1.55 program (Excoffier, 1993) were generated using AMOVA-PREP (Miller, 1998). Both the entire dataset of all 14 populations and a reduced dataset that did not include the two populations of presumed recent origin (*E5 and *E6; Table 1) were analysed in separate AMOVAs. All significance tests used 999 permutations.
In order to avoid potential bias caused by populations *E5 and *E6, the AMOVA based on 12 populations was used for the partitioning of genetic variances among regions, among populations within regions and within populations. Overall ΦST, which is the correlation of random RAPD phenotypes within populations relative to that of random pairs drawn from all populations (Excoffier et al., 1992), and which is analogous to FST, was also obtained from AMOVA 1.55.
AMOVA on the whole dataset generated pairwise genetic differentiations (ΦST) with significance levels among all 14 populations. A Mantel test with 999 permutations was performed to test whether the matrix of genetic differentiations among populations was correlated with the matrix of geographical distances using NTSYS-PC (Rohlf, 1998).
Various estimators of genetic diversity within populations have previously been applied to dominant genetic marker data. We compared: (i) the molecular variance (AMOVA sum of squares divided by N − 1, where N is the sample size per population; Fischer & Matthies, 1998); (ii) the Simpson index adjusted for finite sample size (D=1 − Σ {[ni(ni – 1)]/[N(N – 1)]}, where ni is the number of individuals with RAPD multiband phenotype i within a population); (iii) the Shannon index based on RAPD multiband phenotypes (H=− Σpilog2pi, using ni/N as an estimate of pi); and (iv) the Shannon index based on relative frequencies of polymorphic RAPD markers. The four estimators were all normally distributed according to Kolmogorov–Smirnov tests and highly correlated to each other (Pearson correlation coefficients, r > 0.92, P < 0.01 in all cases). Molecular variance was chosen for subsequent analyses of RAPD variation within populations.
Heteroscedasticities of molecular variance among populations and among regions were tested with Bartlett tests implemented in the AMOVA 1.55 program. Pairwise Bartlett tests also were used to evaluate whether pairs of populations exhibited different amounts of RAPD variation. The relationship between molecular variance and present-day population size was studied with a Spearman rank correlation coefficient.
Using the nine populations with known historical population sizes (Table 1), we tested whether historical bottlenecks had a significant effect on present-day molecular variance (two groups of populations with or without bottlenecks; Mann–Whitney U-test). The number of rare RAPD markers with a frequency of less than 0.1 in the whole dataset was determined per population (Table 1). The frequencies of these RAPD fragments were assumed to represent estimators of the frequencies of rare alleles, discounting their actual number at the locus in a genotype. The effect of recent genetic bottlenecks on rare RAPD markers within populations was tested with a U-test.
Possible effects of random sampling were studied by comparing two groups of populations: (i) eight populations of less than 150 individuals or with historical bottlenecks; (ii) six populations of more than 300 individuals and without bottlenecks (Table 1). In order to test for a difference in population divergence between the two groups, a U-test was performed on the pairwise genetic differentiations (ΦST) among populations within groups. To test for a difference in frequency deviations of geographically widespread RAPD markers, the deviation of their relative frequency within population from their relative frequency in the whole data set was calculated. The average deviations of marker frequencies (AD) within populations were then tested for a significant difference between the two groups (U-test). Standard statistical tests were performed on SPSS (SPSS, 1998).
Results
RAPD diversity
Screening 73 single primers with 12 individuals, each from a different D. cristata population and eight primer combinations with 10 individuals, yielded 361 scorable RAPD fragments, seven (1.9%) of which were polymorphic. When running all 280 individuals, primers OPA-18, OPC-15, OPD-01, OPD-03, OPD-08 and the pair OPA-20/OPB-04 produced nine polymorphic marker fragments ranging between 500 and 1750 bp in length. Only primer OPA-18 revealed more than one polymorphic marker; primer OPD-01 reproduced one of the polymorphisms of primer OPA-18 and was excluded from analysis. All further analyses were therefore based on eight independent polymorphic RAPD markers.
Relative frequencies of the presence of these eight marker fragments ranged from 0.03 to 0.36 in the whole dataset; four rare markers had frequencies of less than 0.10. None of the 14 studied populations of D. cristata was monomorphic for all RAPD markers. Fragment OPA-18.500 was restricted to populations W3 and W4 and fixed in these two populations. Fragment OPD-03.1200 was restricted to populations B1, B2 and B4 and fixed in populations B1 and B2; fragment OPD-08.1050 was restricted to populations B2 and B4. The other five RAPD markers were geographically widespread, including the four rare ones.
The 280 individuals of D. cristata comprised 27 RAPD multiband phenotypes (Appendix), and between two and eight different phenotypes were detected within populations. Three phenotypes occurred in all three investigated regions, and three were found in two regions. Six phenotypes were restricted to particular regions, and 15 phenotypes were restricted to particular populations. All phenotypes sampled in populations W1, W2, E1, E2, E3 and E4 occurred in at least two regions. About 40% of the sampled individuals had Phenotype 1, characterized by the absence of all polymorphic marker bands (Appendix). Most of the 27 RAPD phenotypes were rare, 14 of them occurring only one to three times in the whole dataset (Appendix).
Variance partitioning and genetic differentiation
In the AMOVA on 12 D. cristata populations, variation in RAPD banding patterns was significant among regions, among populations within regions and within populations (P < 0.001 in all cases; Table 2). The greatest amount — approximately half — of the total variance was found within populations, another third was attributed to variation among populations within regions, while the remaining 15% was attributed to variation among regions (Table 2; similar results were obtained from the AMOVA on all 14 populations; data not shown). The overall ΦST-value was 0.49 (P < 0.001), showing that about 50% of total genetic variation was found among populations.
Seventy-two of the 91 pairwise genetic differentiations (ΦST) between the 14 populations were highly significant (P < 0.001), and their values ranged from 0.08 to 0.77. No significant genetic divergence was found for three pairs of populations with historical bottlenecks (E3*E6, *E5*E6, *E5W1; Table 1). Genetic divergence of populations was not significantly correlated with geographical distance (Mantel test, rm=0.168, P > 0.1). Pairwise genetic differentiations among small populations and populations with recent historical bottlenecks were not significantly different from those among large populations (U-test, P > 0.9).
Genetic variation within populations
Molecular variance was significantly different among the 14 populations of D. cristata and among the three regions (Bartlett tests, P < 0.001 in both cases). Significant differences of molecular variance within populations were found in 73 out of 91 pairwise Bartlett tests (P < 0.001 in 72 cases, P < 0.05 in one case). The amount of molecular variance was not significantly different in all six pairs of populations with historical bottlenecks (Table 1). Only two of the comparisons of populations with and without historical bottlenecks showed nonsignificant differences of molecular variance (E2*E6, W1E2; Table 1). Mean molecular variance per population within region was highest in Baden-Württemberg and lowest in eastern Switzerland, i.e. the central region of the study area (Table 1).
No significant relationship between molecular variance and population size was found in all 14 populations of D. cristata (Spearman correlation coefficient, rs=−0.174, P > 0.5; Table 1, Fig. 2), nor in the nine populations with known history (rs=0.288, P > 0.4). However, molecular variance was significantly lower in populations with recent historical bottlenecks than in populations without bottlenecks (U-test, P < 0.02; Table 1, Fig. 3). The number of rare RAPD markers present was not significantly different in bottlenecked populations compared to populations without bottlenecks (U-test, P > 0.5; Table 1).
Mean frequencies of the present rare markers within populations were increased in populations *E5 and *E6 and highest in the two smallest populations B2 and B3 (Table 1). Considering the five geographically widespread RAPD markers, the average deviation of their relative frequencies (AD) was significantly higher in small and/or recently bottlenecked populations [mean= 0.14 ± 0.02 (SE)] compared to large populations without bottlenecks [mean=0.06 ± 0.01; U-test, P < 0.01; Table 1].
Discussion
Extraordinarily low RAPD diversity
Very low genetic diversity was found in 14 populations of D. cristata. Comparison of RAPD diversities of species investigated in different studies is delicate because of varying primer screening strategies and sample sizes. However, RAPDs usually provide high numbers of polymorphic markers (Bartish et al., 1999), and a few primers are often sufficient to distinguish most of the sampled individuals (Fischer & Matthies, 1998; Gugerli et al., 1999). In D. cristata, after a thorough screening for variable markers, only eight polymorphic RAPD markers and 27 multiband phenotypes were found in 280 individuals. Low RAPD diversity has also been detected in D. remota within its whole distribution range, which has been interpreted as evidence for a single origin of this apomictic, triploid fern species (Schneller et al., 1998). A mainly monomorphic RAPD profile has been reported for Pinus resinosa, which was attributed to an ancient bottleneck during glaciation (Mosseler et al., 1992).
Only a small area at the border of the distribution of D. cristata was sampled in the present study. However, our RAPD results were in accordance with very low allozyme polymorphism reported for D. cristata in Europe (Güldenpenning, 1994; J.C. Vogel, Natural History Museum, London, unpubl. data). An explanation for the extraordinarily low genetic diversity in D. cristata might be a single or very few colonization events of Central Europe, or even a single or very few origins of this allotetraploid species, accompanied by considerable inbreeding. The most convincing example of a widespread allopolyploid fern species with a single origin, as deduced from allozyme profiles, is D. celsa (Werth, 1991). This species shares with D. cristata one of its parental diploids, the North American species D. ludoviciana (Werth, 1991). An ancient bottleneck during speciation or colonization history of D. cristata would suggest that the detected RAPD diversity resulted primarily from subsequent mutations, which might also account for the right-skewed frequency distribution of the polymorphic RAPD markers.
The origin of allopolyploids by rare hybrid-polyploidization implements the ability of selfing, and Masuyama & Watano (1990) assumed that a trend for selfing exists in polyploid pteridophytes. In diploid homosporous ferns, intragametophytic selfing produces completely homozygous sporophytes in a single generation. The low number of RAPD multiband phenotypes found in D. cristata may point to high selfing rates.
High population differentiation
High genetic differentiation among populations was found in D. cristata, which exhibited almost 50% of total genetic variation among populations. However, when interpreting RAPD phenotype diversity in D. cristata, one needs to remember the large number of monomorphic RAPD fragments not taken into account in the analysis. Yet extensive screening, tested quality of marker fragments and large sample size still justified a conventional analysis of RAPD data in the present study.
Outcrossing, wind-pollinated and long-lived seed plants maintain most genetic variation within populations, while mainly selfing, short-lived species harbour comparatively higher variation among populations (Hamrick & Godt, 1989; Bartish et al., 1999). The proportion of among-population variation in D. cristata, as measured by RAPDs, was similar to that reported for selfing seed plants based on allozyme diversity (51% on average; Hamrick & Godt, 1989). However, homosporous ferns possess great potential for long-distance spore dispersal (Cousens, 1988), and Soltis & Soltis (1990) argued that the mating system might be of minor importance in determining genetic structures of fern species. Genetic homogeneity among populations was accordingly found in four outcrossing, diploid ferns (FST between 0.02 and 0.11), but considerable interpopulational differentiation (FST=0.23) has been reported for D. expansa, which has a mixed mating system (Soltis & Soltis, 1990). A high proportion of genetic variation among populations was found in the three tetraploid, predominantly selfing ferns, Asplenium septentrionale, A. ruta-muraria and Polypodium vulgare, by Schneller & Holderegger (1996). Intragametophytic selfing in these three species was supposed to be adaptive with regard to single spore colonization of naturally isolated and restricted suitable habitat patches. In summary, the high ΦST-value of 0.49 found in D. cristata indicates comparatively high population differentiation and may provide further evidence for substantial selfing rates in populations of this fern species. Nevertheless, in the multiband phenotypes of populations comprising geographically restricted RAPD fragments, these markers were combined freely with widespread markers, which may provide evidence for occasional recombination in D. cristata.
The missing correlation between genetic divergence and geographical distance and the high population differentiation both indicated limited interpopulational gene flow in D. cristata, despite the potential for spore dispersal over long distances (see above). The absence of gene flow as an equalizing force allowed us to investigate effects of random evolutionary processes on the genetic diversity within populations of D. cristata.
The influence of population history on genetic variation
Population sizes did not explain the differences in molecular variances of the studied populations of D. cristata; genetic variation was not positively correlated with population size (Fig. 2). There was even a slightly negative relationship between the two traits, as the two smallest populations, B2 and B3, showed the highest genetic variation (Table 1). Additionally, the genetic variation of large populations might also have been slightly underestimated in the present study, because only 20 individuals were sampled per population.
Present-day genetic variation of D. cristata populations could be explained by events in recent population history. Historical population bottlenecks were accompanied by serious losses of genetic variation. Molecular variance within bottlenecked populations was reduced by about 40% in comparison with populations without bottlenecks (Table 1, Fig. 3). A similarly genetic diversity has been reported for a reintroduced population of the allotetraploid Argyroxiphium sandwicense ssp. sandwicense (Friar et al., 2000). The four bottlenecked populations of D. cristata comprised substantially fewer rare RAPD phenotypes than most of the other studied populations. However, the theoretical predictions of predominant elimination of rare markers during genetic bottlenecks (Nei et al., 1975) could not be confirmed with RAPD data in D. cristata (Table 1), but has been reported for A. sandwicense ssp. sandwicense (Robichaux et al., 1997). Two populations of D. cristata of presumed recent origin, *E5 and *E6, had similar genetic variation to populations with reported severe population bottlenecks. Low molecular variance and a total lack of rare markers in population B1 suggested a genetic bottleneck in its recent history as well, but no indication for this was found in the floristic literature on southern Germany.
The following discussion of the relationship between different categories of population history and present-day genetic variation in D. cristata has two shortcomings. First, there were only a few cases per category, and, second, populations of D. cristata might have experienced changes in size even before they were floristically investigated. The formerly small populations E1 and E2, which have recently increased in size, showed intermediate values of molecular variance (Table 1). High genetic variation was still found in small remnants of formerly large populations (W2, W4; Table 1). Thus, a distinct reduction in population size to about 60 individuals does not seem to reduce genetic variation severely in the long-lived D. cristata, at least in a short period of about 50 years. This was in accordance with the finding that no substructuring of genetic variation within populations of D. cristata occurred (data not shown). Unfortunately, there was only one presently large population, E4, known to have been large also in the past, which exhibited low genetic variation (Table 1). This population is isolated geographically in the pre-alpine Rhine valley (Fig. 1), which might account for its low genetic variation. Ellstrand & Elam (1993) opined that biologists are more likely to report census numbers for small populations, because they are easier to count; a habit that undermines our ability to reconstruct population histories. In fact, it was not possible to evaluate the historical sizes of the presently large populations B4 and W3, although their locations were already recorded at the end of the 19th century (Willbold, 1995; Landergott et al., 2000). These two populations were not isolated (Fig. 1) and showed considerable genetic diversity (Table 1). They thus would exhibit the genetic profiles of unrestricted populations.
Random sampling effects are expected in populations with recent genetic bottlenecks and/or in small populations. Genetic differentiation among such populations was not increased compared to that among large populations of D. cristata. This was in contrast with results on Salvia pratensis and Scabiosa columbaria, where the coefficient of genetic differentiation (GST) was substantially higher among small than among large populations (van Treuren et al., 1991). Rare RAPD markers in D. cristata reached high frequencies in the two smallest populations, B2 and B3, and in recently founded populations, *E5 and *E6 (Table 1). High frequencies of rare alleles were exclusively observed in small populations of Salvia pratensis and Scabiosa columbaria, supposed to be caused by genetic drift (van Treuren et al., 1991). Under genetic drift, all alleles — not only the rare ones — are affected by random sampling, and the probability of an increase in frequency is the same as that of a decrease, regardless to allele frequency (Hartl & Clark, 1997). The average deviation of relative frequencies of the five geographically widespread RAPD markers was significantly higher in small and bottlenecked populations of D. cristata than in large populations (Table 1). This can be interpreted as evidence for random genetic drift. The highest average deviations were found in the two smallest populations, B2 and B3, each with a population size of about 25 individuals. A substantially increased deviation was also detected in population W4, with 110 individuals. Although comparatively high levels of genetic variation were still maintained in these latter populations, they might be prone to stochastic loss of genetic diversity in the future. The lowest average deviation of widespread marker frequencies was found in population B4, a probably old and large population (see above). Furthermore, the relatively large number of neighbouring populations of D. cristata increased the probability of interpopulational gene flow in the region of Baden-Württemberg (Fig. 1).
The relationship between genetic diversity and population viability is controversial (see, e.g., Fischer & Matthies, 1998; Schmidt & Jensen, 2000). Ecological factors might be more important in this context than genetic factors (Ellstrand & Elam, 1993). Viable populations with complete allozyme uniformity have been reported for predominantly inbreeding, polyploid fern species (Schneller & Holderegger, 1996). No fitness parameters were measured in D. cristata. Nevertheless, recently increasing population sizes and a comparatively high spore production observed in the region of eastern Switzerland (Landergott et al., 2000) might indicate that, in the short term, suitable environmental conditions are more important for population viability in this fern species than genetic variation, which was lowest in that very region (Table 1).
Conclusions and implications for conservation
Populations of D. cristata exhibited two somewhat conflicting genetic features, namely the very low overall genetic diversity and the comparatively strong population differentiation. In conservation biology, the number of multilocus genotypes present in a population is supposed to be a more important measurement of genetic diversity than the number of single markers (Schneller & Holderegger, 1996). Without knowledge of possible adaptive differences among populations of D. cristata, the best conservation strategy would be the maintenance of genetic differences and the priority preservation of populations with comparatively high genetic diversity. In western Switzerland, where D. cristata is the focus of a local conservation project (Kozlowski, 1999), populations W1 and W2 were genetically different from populations W3 and W4. The maintenance of regional genetic diversity would therefore require the conservation of both pairs of populations. High genetic variation was maintained in the remnants of the formerly large populations W2 and W4. However, such small populations might be prone to future loss of genetic diversity through random genetic drift (see above).
Substantial historical population dynamics should be expected in newly rare, threatened species, because increasing rarity is often caused by distinct habitat changes resulting from human activities. In fact, recent population histories better explained present-day genetic diversity in populations of D. cristata than did actual population sizes. Serious reductions of genetic variation in natural populations of this long-lived, polyploid fern species were the result of severe recent historical bottlenecks. Small remnants of formerly large populations of D. cristata exhibited higher genetic diversity than did large, but formerly small populations. Nevertheless, there was evidence for random genetic drift acting upon small populations of D. cristata. Hence, the present study exemplifies how an extensive survey of floristic data aids the interpretation of genetic data and stresses the importance of population history for population genetics and conservation biology.
References
Bachmann, K. (1997). Nuclear DNA markers in plant biosystematic research. Opera Bot, 132: 137–148.
Barrett, S. C. H. and Kohn, J. R. (1991). Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk, D. A. and Holsinger, K. E. (eds) Genetics and Conservation of Rare Plants, pp. 3–30. Oxford University Press, Oxford.
Bartish, I. V., Jeppsson, N. and Nybom, H. (1999). Population genetic structure in the dioecious pioneer plant species Hippophaë rhamnoides investigated by random amplified polymorphic DNA (RAPD) markers. Mol Ecol, 8: 791–802
Cousens, M. I. (1988). Reproductive strategies of pteridophytes. In: Lovett Doust, J. and Lovett Doust, L. (eds) Plant Reproductive Ecology: Patterns and Strategies, pp. 307–328. Oxford University Press, Oxford.
Dostal, J., Reichstein, T. and Fraser-Jenkins, C. (1984). Aspidiaceae. In: Kramer, K. U. (ed.) Hegi G. Illustrierte Flora von Mitteleuropa, Vol. II(1), pp. 132–187. Parey, Berlin.
Ellstrand, N. C. and Elam, D. R. (1993). Population genetic consequences of small population size: implications for plant conservation. Ann Rev Ecol Syst, 24: 217–242.
Excoffier, L. (1993). AMOVA. Analysis of Molecular Variance, Release 1.55. Genetics and Biometry Laboratory, University of Geneva, Geneva.
Excoffier, L., Smouse, P. E. and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial-DNA restriction data. Genetics, 131: 479–491.
Fischer, M. and Matthies, D. (1998). RAPD variation in relation to population size and plant fitness in the rare Gentianella germanica (Gentianaceae). Am J Bot, 85: 811–819.
Friar, E. A., Ladoux, T., Roalson, E. H. and Robichaux, R. H. (2000). Microsatellite analysis of a population crash and bottleneck in the Mauna Kea silversword, Argyroxiphium sandwicense ssp. sandwicense (Asteraceae), and its implications for reintroduction. Mol Ecol, 9: 2027–2034
Godt, M. J. W., Johnson, B. R. and Hamrick, J. L. (1996). Genetic diversity and population size in four rare southern Appalachian plant species. Conserv Biol, 10: 796–805.
Gugerli, F., Eichenberger, K. and Schneller, J. J. (1999). Promiscuity in populations of the cushion plant Saxifraga oppositifolia in the Swiss Alps as inferred from random amplified polymorphic DNA (RAPD). Mol Ecol, 8: 453–461.
Güldenpenning, S. (1994). Isoenzymatische Untersuchungen an ausgewählten Farnen Deutschlands. Diploma Thesis, Ruhr-University, Bochum.
Hamrick, J. L. and Godt, M. J. W. (1989). Allozyme diversity in plant species. In: Brown, H. D., Clegg, M. T., Kahler, A. L. and Weir, B. S. (eds) Plant Population Genetics, Breeding, and Genetic Resources, pp. 43–63. Sinauer, Sunderland, MA.
Hartl, D. L. and Clark, A. G. (1997). Principles of Population Genetics. Sinauer, Sunderland, MA.
Kozlowski, G. (1999). Ein Arten- und Biotopschutzprojekt des Botanischen Gartens Freiburg: Bedrohte und seltene Pflanzenarten in den Mösern von Düdingen und Sâles (Kt. Freiburg). Schwerpunkt: Der Kammfarn (Dryopteris cristata (L.) Gray) und die Pflege seiner Lebensräume. Bull Soc Frib Sci Nat, 88: 58–70.
Lande, R. and Shannon, S. (1996). The role of genetic variation in adaptation and population persistence in a changing environment. Evolution, 50: 434–437.
Landergott, U., Schneller, J. J., Holderegger, R. and Kozlowski, G. (2000). Populationsgeschichte des seltenen Kammfarns (Dryopteris cristata) in der Schweiz. Bot Helv, 110: 151–170.
Lutz, E., Schneller, J. J. and Holderegger, R. (2000). Understanding population history for conservation purposes: population genetics of Saxifraga aizoides (Saxifragaceae) in the lowlands and lower mountains north of the Alps. Am J Bot, 87: 583–590.
Masuyama, S. and Watano, Y. (1990). Trends for inbreeding in polyploid pteridophytes. Pl Sp Biol, 5: 13–17.
Miller, M. P. (1998). AMOVA-PREP. A Program for the Preparation of AMOVA Input Files from Dominant-Marker Raw Data, Release 1.01. Department of Biological Sciences, Northern Arizona University, Flagstaff, AZ.
Mosseler, A., Egger, K. N. and Hughes, G. A. (1992). Low levels of genetic diversity in red pine confirmed by random amplified polymorphic DNA markers. Can J For Res, 22: 1332–1337.
Nei, M., Maruyama, T. and Charkaborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution, 29: 1–10.
Robichaux, R. H., Friar, E. A. and Mount, D. W. (1997). Molecular genetic consequences of a population bottleneck associated with reintroduction of the Mauna Kea silversword (Argyroxiphium sandwicense ssp. sandwicense (Asteraceae). Conserv Biol, 11: 1140–1146.
Rohlf, F. J. (1998). NTSYS-PC. Numerical Taxonomy and Multivariate Analysis System, Release 2.02i. Exeter Software, Setauket, NY, USA.
Schmidt, K. and Jensen, K. (2000). Genetic structure and AFLP variation of remnant populations in the rare plant Pedicularis palustris (Scrophulariaceae) and its relation to population size and reproductive components. Am J Bot, 87: 678–689.
Schneller, J. J. and Holderegger, R. (1996). Genetic variation in small, isolated fern populations. J Veg Sci, 7: 113–120.
Schneller, J. J., Holderegger, R., Gugerli, F., Eichenberger, K. and Lutz, E. (1998). Patterns of genetic variation detected by RAPDs suggest a single origin with subsequent mutations and long-distance dispersal in the apomictic fern Dryopteris remota (Dryopteridaceae). Am J Bot, 85: 1038–1042.
Schwaegerle, K. E. and Schaal, B. A. (1979). Genetic variability and founder effect in the pitcher plant Sarracenia purpurea L. Evolution, 33: 1210–1218.
Skroch, P. and Nienhuis, J. (1995). Impact of scoring error and reproducibility of RAPD data on RAPD based estimates of genetic distance. Theor Appl Genet, 91: 1086–1091.
Soltis, P. S. and Soltis, D. E. (1990). Genetic variation within and among populations of ferns. Am Fern J, 80: 161–172.
SPSS (1998). SPSS FOR WINDOWS, Release 9.0. SPSS Inc., Chicago, IL.
Van Treuren, R., Bijlsma, R., Van Delden, W. and Ouborg, N. J. (1991). The significance of genetic erosion in the process of extinction. I. Genetic differentiation in Salvia pratensis and Scabiosa columbaria in relation to population size. Heredity, 66: 181–189.
Walker, S. (1961). Cytogenetic studies in the Dryopteris spinulosa complex. II. Am J Bot, 48: 607–614.
Walker, S. (1969). Identification of a diploid ancestral genome in the Dryopteris spinulosa complex. Brit Fern Gaz, 10: 97–99.
Weising, K., Nybom, H., Wolff, K. and Meyer, W. (1995). DNA Fingerprinting in Plants and Fungi. CRC Press, Boca Raton, FL.
Werth, C. R. (1989). Isozyme evidence on the origin of Dryopteris cristata and D. carthusiana. Am J Bot, 76(Suppl.)208–208.
Werth, C. R. (1991). Isozyme studies on the Dryopteris ‘spinulosa’ complex. I. The origin of the Log Fern Dryopteris celsa. Syst Bot, 16, 446–461.
Willbold, E. (1995). Der Kammfarn (Dryopteris cristata (L.) A. Gray) in Württemberg. Jh Ges Naturk Württemberg, 151: 429–437.
Young, A., Boyle, T. and Brown, T. (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol, 11: 413–418
Acknowledgements
We thank Elmar Willbold for providing information on populations of D. cristata in southern Germany, Johannes Vogel for insight into unpublished data and the Eugen Baumann-Foundation for financial support for travelling expenses. Laboratory costs were funded by the Georges and Antoine Claraz-Foundation. We gratefully acknowledge Daniela Lang for help in the laboratory and Elena Conti, Austin Mast, Ivana Stehlik and two anonymous reviewers for helpful comments on earlier versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Landergott, U., Holderegger, R., Kozlowski, G. et al. Historical bottlenecks decrease genetic diversity in natural populations of Dryopteris cristata. Heredity 87, 344–355 (2001). https://doi.org/10.1046/j.1365-2540.2001.00912.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2001.00912.x
Keywords
This article is cited by
-
The impact of selection on population genetic structure in the clam Meretrix petechialis revealed by microsatellite markers
Genetica (2016)
-
Elucidating Genetic Diversity among Sour Orange Rootstocks: a Comparative Study of the Efficiency of RAPD and SSR Markers
Applied Biochemistry and Biotechnology (2015)
-
Assessment of genetic diversity in Pithecellobium dulce (Roxb.) Benth. germplasm using RAPD and ISSR markers
Trees (2015)
-
Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers
Molecular Biology Reports (2015)
-
The structural and functional connectivity of the grassland plant Lychnis flos-cuculi
Heredity (2014)