Abstract
The colonization dynamics of the black truffle in an artificial field were assessed through analyses of microsatellite and RAPD markers. The truffle field was composed of three tree species and mycelial inoculum of three different origins, and was monitored for the first three years of truffle production. We found very low levels of genetic diversity. Isolation by distance was detected only at the between-tree level. This could be interpreted as local colonization around each tree facilitated by the presence of the tree root system. At the larger spatial scale of the European range, the absence of isolation by distance corroborates the hypothesis of an impact of glaciation on genetic variation, followed by rapid postglaciation demographic expansion. In addition, genetic variation of harvested truffles was explained by neither inoculation origin, nor tree species. Our study questions the real impact of man-made inoculation of tree root systems with fungal mycelia.
Similar content being viewed by others
Introduction
The black truffle (Tuber melanosporum Vitt.) is a hypogeous ectomycorrhizal ascomycete mainly found in France, Italy and Spain (Callot, 1999), and highly appreciated for its remarkable organoleptic properties (taste and odour). Haploid mycelium can be found in the soil, usually in contact with or near to tree root systems. Fruiting bodies (i.e. ascocarps) result from the fusion of two haploid mycelia, and are therefore supposedly diploid (Moore-Landecker, 1996). They are composed of conjunctive tissue (the gleba) that surrounds haploid spores. These spores are produced by meiosis after karyogamy.
The black truffle naturally occurs on calcareous soils, in association with the root systems of trees, most often oaks (Quercus spp.) and hazelnuts (Corylus avellana). Since the 1960s, attempts have been made to create favourable artificial habitats to grow and monitor truffles, resulting in managed fields of planted trees.
Traditionally, a truffle field is created by sowing oak acorns and relying on the presence of truffle mycelium in the soil to interact with the developing tree root system. Under such conditions the first truffles are usually harvested about 6–8 years after sowing. Alternatively, it is now possible to inoculate the roots of young trees with specific mycelia before planting these trees directly in the soil (Chevalier & Grente, 1979). Truffles are harvested earlier (3–4 years) under these conditions (Chevalier & Dupré, 1988) and can even be grown in areas where the mycelium did not previously occur (e.g. New Zealand and Tasmania; Callot, 1999). However, when T. melanosporum mycelia or spores already exist in the soil (mainly in France, Italy and Spain), the usefulness of planting inoculated trees is unclear. In particular, it has not been established whether the resulting truffles originated from the artificially generated mycorrhiza or from formerly present local (and potentially competitive) mycelia. In other words, when a black truffle is harvested around an artificially inoculated tree, there is no simple way of determining whether the former has arisen from the inoculated genetic entity (hereafter called ‘clone’, due to its propagation by mycelial growth). Sometimes trees inoculated with T. melanosporum produce truffles of a distinctly different species (e.g. T. brumale or T. uncinatum; Callot, 1999). In addition, the dynamics of spatial colonization are currently unknown: do they correspond to an individual mycelium developing around a particular tree and subsequently spreading to nearby trees, or are there as many independent starting points as producing trees?
We begin by quantifying the genetic diversity of a truffle field at different spatial scales. Second, we explore spatial and temporal colonization patterns through sampling and genetic analysis of the truffle field during its first years of production. We developed microsatellite and RAPD markers to assess the genetic heterogeneity at the fine-grained level of the tree root system and between trees in the same field.
Materials and methods
Data sampling and DNA extraction
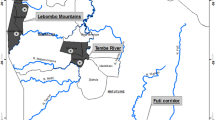
The truffle field studied was located in Viols-le-Fort, near Montpellier, southern France. Holm and Downy Oaks (Quercus ilex and Q. pubescens, respectively) and hazelnut trees (Corylus avellana) were planted between 1989 and 1991. All trees were previously inoculated with Tuber melanosporum. The inoculum had three different origins: INRA (French National Institute of Agronomic Research, 24 trees), a local nursery (eight trees), and the owner of the truffle field (11 trees). The truffle field was monitored during the first three years of production (1995–97), and all detected truffles were sampled using a trained dog (Fig. 1). The location of each truffle ascocarp around its associated tree was recorded. Truffles collected in various areas of France and Italy (previously analysed in Bertault et al., 1998) were added to the present data set for additional testing of the observed relationships at a wider geographical range.
Map of the truffle field. Three tree species were planted: hazelnuts (square), Holm oaks (circle), and Downy oaks (triangle). All trees were inoculated with either mycelium coming from INRA (squared trees), the tree nursery (circled trees), or the truffle field owner (otherwise). Filled symbols indicate trees that produced at least one harvested ascocarp during the sampling period.
Each ascocarp was carefully washed, flame-sterilized, peeled and the inner part was cut in small pieces under sterile conditions, and then freeze-dried. DNA was extracted using a phenol–chloroform protocol (Lee et al., 1988), and stored in 200 μL of a Tris–EDTA 0.1× buffer at −20°C. This DNA solution was diluted 1/100 for PCR reactions.
Genetic marker isolation
RAPD RAPD primers from the OPB and OPF RAPD 10-mer priming kits (Operon Technologies) were used. PCR reactions were performed according to modified conditions described in Williams et al. (1990) and Welsh & McClelland (1990): after an initial denaturation step of 4 min at 95°C, only 30 cycles were performed, each of 1 min at 95°C, 1 min at 36°C and 1.5 min at 72°C. The final step was 72°C for 15 min.
Microsatellites. Two different protocols for genomic library construction and screening were employed. The first one began with the digestion of 100–150 μg DNA using 50 units of Sau3A, followed by isolation of the 300–600 bp DNA fragment fraction. Ligation to pUC19 plasmid vector was followed by heat-shock transfection into XL1-Blue MRF′ competent E. coli bacteria (Stratagene). Colour-discriminated, insert-containing cells were isolated and plasmid DNA was extracted through a ‘miniprep’ protocol (according to manufacturer’s recommendations). Screening of the libraries was done with direct PCR amplification on bacterial colonies as described in Güssow & Clackson (1989) followed by hybridization with 32P radio-labelled probes. The second microsatellite isolation protocol included a microsatellite DNA enrichment step, closely following Kijas et al. (1994). Library screening was done using PCR reactions according to Waldbieser (1995). For both genomic library construction protocols, positive clones were sequenced by cycle-sequencing using 33P-labelled ddNTP.
Genetic analyses
Each variable RAPD band was considered to be a putative locus and coded as present/absent. The combination of these RAPD loci and the variable microsatellite loci yielded the basic data set. Heterozygote deficits at the variable microsatellite loci were tested using an exact U-score test (Rousset & Raymond, 1995). Independence between loci was tested using a Fisher exact test on a R × C contingency table. Geographical co-ordinates of each ascocarp were calculated: (1) for the 147 truffles harvested in the truffle field; and (2) with 50 additional truffles sampled elsewhere in France and Italy. These co-ordinates were used to perform isolation-by-distance analyses both at the local and global spatial scales (according to Rousset, 1997). Except for isolation by distance, all computations were performed using version 3.2 of the software package GENEPOP (Raymond & Rousset, 1995). Methods for the analysis of isolation by distance on pairs of diploid individuals are described in Rousset (2000). These methods require estimation of gene diversities within individuals (heterozygosities), or, alternatively, other information such as gene diversities among individuals competing for a given location. Because the latter information was not available, only the numerator of Rousset’s (2000) â statistic (estimating the ratio (Qw − Qr)/(1 − Qw), where Qr is the probability of identity of genes at geographical distance r and Qw is the probability of identity of two genes within an individual) could be directly estimated. The genetic distance index employed was the proportion of different single-locus genotypes between two truffles. This statistic was used for qualitative analyses of patterns of isolation by distance, requiring a slightly modified version of programs described in Rousset (2000) and included in version 3.2 of GENEPOP. For analyses among trees, a matrix of differences between trees was constructed from the average values of differences between pairs of truffles for each pair of trees.
Results
Isolating genetic markers
RAPD. Twenty-five primers were tested on T. melanosporum DNA, nine of which (OPF-04, -05, -06, -09, -11, -13, -14, -16 and OPB-02) showed clear and reproducible amplification patterns. These nine primers were assayed on a subset of 19 truffle ascocarps from various geographical origins in France and Italy. The four primers (OPB-02, OPF-11, -13 and -14) displaying the highest band variability on the truffle subset were used to analyse all other truffles. This procedure provided six polymorphic bands (two for OPF-13 and OPF-14, and only one for OPB-02 and OPF-11), corresponding to six presumptive loci with two alleles (presence/absence of a band). RAPD patterns also displayed a large number of monomorphic bands, present in all ascocarps (details not shown, see Fig. 2 for example).
RAPD banding patterns in (a) Tuber melanosporum, compared to (b) T. aestivum. Patterns were obtained with primer OPF-14; M, molecular weight marker; C, negative PCR control; numbers refer to different sampled populations (note there is no correspondence between population numbers of T. melanosporum and T. aestivum).
Microsatellites. The standard protocol for microsatellite isolation was performed twice. The first library was screened for seven microsatellite classes ((GACA)n, (TC)n, (TG)n, (CAC)n, (CTC)n, (CTAT)n and (TGTA)n), yielding 28 positive clones out of 720 clones (3.9%). The second library was screened for eight microsatellite classes (the previous seven except (GACA)n, plus (AAT)n and (AAG)n), yielding 84 positive clones out of 2750 clones (3.1%). Finally, the enriched library, screened with two motifs ((TG)n and (TC)n), gave 28 positive clones out of 1015 clones (2.8%). It is noteworthy that the enriched library did not yield better results than the unenriched libraries (Chi-squared test, P=0.40).
Eleven clones (one, five, and five for the three consecutive libraries, respectively) were useful microsatellite loci and were tested for polymorphism on a sample of 38 ascocarps of various origins that showed different RAPD banding patterns (Table 1). Out of these 11 loci, seven were monomorphic and two were polymorphic (F12I and H1b), each displaying only two alleles differing by one repeat unit. Two loci (TM458 and A9V) showed abnormal polymorphisms of 2 and 1 base pairs (bp), respectively. This was not congruent with the 3, 4, or 5 bp length of the repeat units for TM458 and the 2 bp unit for A9V, and was probably due to insertion/deletion mutations in the flanking regions. As these polymorphisms were expected to be much lower than those expected with microsatellite loci, they were not considered further.
Genetic analyses
Only two truffles were found during the first year (1995) of production, followed by 69 in 1996, and 76 in 1997. All these 147 ascocarps were genotyped for the two microsatellite loci and the six presumptive RAPD loci. No heterozygote was identified for any of the two microsatellite loci, indicating significant (P < 10−4) heterozygote deficiency. This is consistent with a rather strict homothallism (i.e. homogamy), or perhaps even selfing. Consequently, we considered each truffle as representing a single gene sampling unit (Table 2).
At the tree level, coexistence of many clones was observed (Fig. 3): over the sampling period, 10 out of the 43 trees that produced at least one ascocarp had more than one associated clone, with an average of 2.6 different clones per tree (mean number of harvested ascocarps per such tree = 6.0). The location of these clones around their associated tree was not related to their genetic composition (global test, Fisher’s method, on all within-tree isolation-by-distance tests, χ2=13.07, d.f.=10, P=0.22).
Over the whole truffle field, there was significant isolation by distance (Mantel test for correlation between the matrices of genetic and geographical distances, 20 000 permutations, P < 10−4, Fig. 4). This effect remained when the data set was split by year (1996: P=0.003; 1997: P < 10−3). It also remained (P < 0.01) when only one ascocarp per multilocus genotype and per tree was considered, or when the truffle contributing to most of the extreme points (in the 0.625 and 0.75 genetic distance categories, the highest two categories, see Fig. 4) in the between-tree group, was removed.
Overall isolation by distance in Tuber melanosporum. The three groups of plots correspond to within trees (distance < 5 m), between trees, within the truffle field (distance < 150 m), and between populations (5 m < distance > 150 m). Regression lines correspond to: within-tree (dotted), between trees (dashed), overall truffle field (dotted-dashed) and whole sampling area (plain).
Genetic differences were not related to the origin of tree inoculation (Mantel test for correlation between the genetic distance matrix and a matrix with ‘0’ for same origin truffles and ‘1’ for truffles of different origins, P=0.65). In other words, two truffles harvested from trees with the same inoculation origin were not more genetically related than two truffles arising from trees with different origins of inoculation. This was consistent with the patterns of isolation by distance found when only pairs of truffles within each origin were compared (Mantel tests, P < 10−4 for INRA trees, P < 10−3 for those planted by the owner of the truffle field, and P=0.20 for the nursery gardener [however, only 12 truffles were analysed in the last case]). In addition, we found no relation between truffle genetic diversity and the tree species around which they were harvested (Mantel test, P=0.67).
Linkage disequilibrium analyses yielded only three significant pairwise locus comparisons out of 28 (using a modification of the Bonferroni sequential test; Hochberg, 1988), suggesting some linkage disequilibrium between certain loci. However, this result was expected, due to geographical structure of the genetic diversity (if the truffles sampled at the tree level result from a single clone, then linkage disequilibrium between genes is expected).
Discussion
Our study reports two particularly striking findings: (1) the low level of genetic variability; and (2) the spatial distribution of this variability.
Low genetic diversity
The low level of genetic variability was observed for both microsatellite and RAPD loci. Only two microsatellite loci out of the 11 isolated were polymorphic, with only two alleles present. In addition, these two alleles diverged by only one repeat unit. Similarly, the number of polymorphic RAPD bands was low compared to the number of monomorphic bands: this low polymorphism was even more apparent when compared to the closely related summer truffle, T. aestivum (Fig. 2). These results indicate that relative to other truffle species, T. melanosporum is genetically depauperate. In particular, T. uncinatum, T. aestivum, T. rufum, T. mesentericum, T. macrosporum, T. borchii and T. excavatum display relatively high levels of genetic diversity based on RAPD primers (Gandeboeuf et al., 1997). Nonetheless, it is interesting that two other European species also show low levels of genetic variability (T. brumale and T. magnatum), although sample sizes were more limited (Gandeboeuf et al., 1997). We suggest that there are two different groups of European truffle of the genus Tuber with regard to their genetic diversities.
The low genetic diversity displayed by T. melanosporum could have two distinct origins: either remains of a pre-bottleneck polymorphism, or new variability created by post-bottleneck mutations. Although the former could not be rejected, the microsatellite data are consistent with the latter. This is because the two alleles at each locus (F12I and H1b, see Table 1) are separated by a single repeat unit, suggesting a recent, single-step mutation event. It is difficult to deduce a date for the bottleneck based on the genetic data alone, but hints from the recent history and the ecology of the black truffle allow some inferences. There is no historical record of any recent T. melanosporum population crash: about 1–3 × 104 kg/year are sold nowadays in France, and sales were even more important at the beginning of the century. Moreover, the high sensitivity of T. melanosporum to cold and frost (Callot, 1999) suggests that a strong bottleneck would have occurred during the last glaciation. This truffle species grows in open habitats, with sparse tree vegetation, meaning that the fungus is susceptible to mortality due to freezing. An important restriction both in range and population size is thus likely to have occurred during the last glaciation (Bertault et al., 1998). This climatic event is thought to have had pronounced effects on the genetic structure and genetic variability of a variety of species (e.g. Highton & Webster, 1976; Schwaegerle & Schaal, 1979; Sage, 1986; Hewitt, 1996; Merilä et al., 1997; Bucklin & Wiebe, 1998).
Microhabitat could also explain the more general pattern of genetic diversity between the two variability groups: all truffle species displaying a low diversity share the same kind of rather sparsely wooded habitat; conversely, the more variable species tend to grow in more forest-like habitats. Such conditions lead to buffered temperatures and could have ensured large enough population sizes to avoid severe genetic bottlenecks.
The very low genetic variability of T. melanosporum possibly reflects that it was on the brink of extinction during the last glaciation. Given individual mortalities and the protected nature of the ascocarp, the persistence of truffles requires some degree of dispersal, and, in particular, requires the attraction of spore vectors via chemical cues. Among truffle species having experienced a strong bottleneck during the last glaciation, those not producing sufficiently intense and elaborate chemical bouquets may have gone extinct. This could explain the intriguing fact that of the three genetically homogeneous truffle species, two (T. melanosporum and T. magnatum) have a strong enough smell to be highly valued by man, a species with notoriously low olfactory abilities.
Isolation by distance
The overall distribution of the genetic diversity is shown in Fig. 4, where three levels are apparent. The first one is the close surroundings of the tree. At this scale, many truffles have the same genotype at all loci. Since ascocarps are thought to represent the product of sexual reproduction, the 100% homozygosity displayed at the microsatellite loci indicates that clones themselves are genetically homogeneous. This suggests that the mode of reproduction is likely to be strict selfing. This particular over-representation of similar combinations of genotypes certainly accounts for the linkage disequilibrium found at some pairs of loci. However, the simultaneous presence of different clones (for all trees producing at least two truffles, mean number of clones = 1.96, max=6, see Fig. 4) around some trees shows that several independent associations with a single tree can occur. In addition, the same clone is able to persist around the same tree: during the two years with sufficient truffle production, 62% (or 10/16) of productive trees displayed the same truffle clone consecutively.
The second level of genetic differentiation is the whole truffle field. Highly significant isolation by distance was found at this scale (Fig. 4), mainly due to the greater differentiation between truffles harvested from different trees than between truffles harvested on the same tree. This indicates that T. melanosporum migrates at a higher rate around a tree than between trees, suggesting, for instance, that migration at this scale is facilitated by the root system.
It is worth noting that no isolation by distance was found when only truffles harvested on different trees were considered. This result could be explained by two hypotheses, depending whether or not the truffle population has reached a migration/drift equilibrium. If the migration/drift equilibrium has been reached, then the absence of isolation by distance indicates that migration usually occurs at a larger scale than the truffle field. Possible spore vectors include the wild boar, known to ingest Tuber ascocarps heavily in southern France and to move over very large distances daily (Genard et al., 1986; Gerard et al., 1991), although other animals could also significantly contribute (e.g. man, see below). If there is no migration/drift equilibrium, then the absence of isolation by distance could be attributed to the direct influence of management of the truffle field on the genetic structure. This could be the result of the practice of randomly distributing crushed truffle (issued from small or overripe unsold truffles) over the whole field and/or the use of truffle inoculum issued from a single truffle field. In the former case, genetic structure is best described by an island model at the scale of the truffle field, and the relevant scale of study is probably above this level. This assumes that the spores sown by the grower germinate and that the harvested truffles do not all result from the artificially inoculated mycorrhiza. In the latter case, the resulting genetic structure should be more related to the origin of these inoculated mycorrhiza than to their geographical location. However, this is not supported by the data, as indicated by the absence of a relation between genotype and inoculum origin. Future studies should distinguish the contribution of these two mechanisms by analysing mycorrhiza directly.
The third level of genetic differentiation is the whole sampling area. At this scale, no relation of isolation by distance was found (Fig. 4). This result is compatible with the hypothesis of a drastic range restriction followed by rapid demographic expansion (Bertault et al., 1998). The absence of isolation by distance displayed by the black truffle over such a large area probably results from both the brevity of the colonization event and the absence of significant recovery of genetic diversity.
It should be emphasized that during the last three decades there have been numerous transfers between artificial truffle fields in Europe. These exchanges consist mainly of young inoculated trees of various origins (France, Spain and Italy), and in principle they could have an impact on genetic structure if the harvested truffles do indeed originate from the inoculated mycelia. However, all sampling reported here (except from the focal truffle field) was conducted on natural sites of the black truffle, thereby reducing this potential bias.
To conclude, most variation in black truffle genotypes occurs between adjacent trees, since it is the largest spatial scale showing a significant pattern of isolation by distance. Our results seriously question the importance of mycorrhizal inoculation as a means of enhancing truffle production, although more data are required to be more conclusive. We suggest that studies focus on what is occurring in the proximity of individual trees rather than between fields, at least during the first years of interaction between the tree and the truffle mycelia.
References
Bertault, G., Raymond, M., Berthomieu, A., Callot, G. et al (1998). Trifling variation in truffles. Nature, 394: 734–734, 10.1038/29428.
Bucklin, A. and Wiebe, P. H. (1998). Low mitochondrial diversity and small effective population sizes of the Copepods Calanus finmarchicus and Nannocalanus minor: possible impact of climatic variation during recent glaciation. J Hered, 89: 383–392.
Callot, G. (1999). La Truffe, la Terre, la Vie. INRA Editions, Paris.
Chevalier, G. and Dupré, C. (1988). Recherche et expérimentation sur la truffe et la trufficulture en France. pp. 157–166. 2° Congresso Internazionale sul Tartufo, Spoleto, Italy.
Chevalier, G. and Grente, J. (1979). Application pratique de la symbiose ectomycorhizienne: production à grande échelle de plants mycorhizés par la truffe. Mushroom Sci, 10: 483–505.
Gandeboeuf, D., Dupré, C., Roeckel-Drevet, P., Nicolas, P. et al (1997). Grouping and identification of Tuber species using RAPD markers. Can J Bot, 75: 35–45.
Genard, M., Lescourret, F. and Durrieu, G. (1986). Mycophagie chez le sanglier et dissemination des spores de champignons hypogés. Gaussenia, 2: 17–23.
Gerard, J. -F., Teillaud, P., Spitz, F., Mauget, R. et al (1991). Le sanglier. Rev d’Ecol, 6 (Suppl), 11–66.
Güssow, D. and Clackson, T. (1989). Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucl Acids Res, 17: 4000–4000.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc, 58: 247–276.
Highton, R. and Webster, T. P. (1976). Geographic protein variation and divergence in populations of the salamander Plethodon cinereus. Evolution, 30: 33–45.
Hochberg, Y. (1988). A sharper Bonferroni procedure for multiple tests of significance. Biometrika, 75: 800–802.
Kijas, J. M. H., Fowler, J. C. S., Garbett, C. A. and Thomas, M. R. (1994). Enrichment of microsatellites from the Citrus genome using biotinylated oligonucleotide sequences bound to streptavidin-coated magnetic particles. BioTechniques, 16: 187–193.
Lee, S. B., Milgroom, M. G. and Taylor, J. W. (1988). A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet Newsl, 35: 23–24.
Merilä, J., Björklund, M. and Baker, A. J. (1997). Historical demography and present day population structure of the greenfinch Carduelis chloris – an analysis of mtDNA control-region sequences. Evolution, 51: 946–956.
Moore-Landecker, E. (1996). Fundamentals of the Fungi. Prentice Hall, Upper Saddle River, NJ.
Raymond, M. and Rousset, F. (1995). GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered, 86: 248–249.
Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145: 1219–1228.
Rousset, F. (2000). Genetic differentiation between individuals. J Evol Biol, 13: 58–62, 10.1046/j.1420-9101.2000.00137.x.
Rousset, F. and Raymond, M. (1995). Testing heterozygote excess and deficiency. Genetics, 140: 1413–1419.
Sage, R. D. (1986). Pleistocene glaciations, fluctuating ranges, and low genetic variability in a large mammal (Ovis dalli). Evolution, 40: 1092–1095.
Schwaegerle, K. and Schaal, B. A. (1979). Genetic variability and founder effect in the pitcher plant Sarracenia purpurea L. Evolution, 33: 1210–1218.
Waldbieser, G. C. (1995). PCR-based identification of AT-rich tri- and tetranucleotide repeat loci in an enriched plasmid library. BioTechniques, 19: 742–744.
Welsh, J. and Mcclelland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucl Acids Res, 18: 7213–7218.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. et al (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res, 18: 6531–6535.
Acknowledgements
We are very grateful to G. Serane for his enthusiastic participation in this study and for the open access to his truffle field. Research was supported in part by DADP-INRA Région Languedoc–Roussillon programme. This is paper no. 2001–048 of the Institut des Sciences de l’Evolution.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bertault, G., Rousset, F., Fernandez, D. et al. Population genetics and dynamics of the black truffle in a man-made truffle field. Heredity 86, 451–458 (2001). https://doi.org/10.1046/j.1365-2540.2001.00855.x
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2001.00855.x
Keywords
This article is cited by
-
Bacterial communities associated with mushrooms in the Qinghai-Tibet Plateau are shaped by soil parameters
International Microbiology (2022)
-
Genotypic diversity of the Asiatic black truffle, Tuber himalayense, collected in spontaneous and highly productive truffle grounds
Mycological Progress (2020)
-
Soil spore bank in Tuber melanosporum: up to 42% of fruitbodies remain unremoved in managed truffle grounds
Mycorrhiza (2019)
-
Certainties and uncertainties about the life cycle of the Périgord black truffle (Tuber melanosporum Vittad.)
Annals of Forest Science (2016)
-
Impact of the competition between mating types on the cultivation of Tuber melanosporum: Romeo and Juliet and the matter of space and time
Mycorrhiza (2014)