Abstract
We cloned and sequenced DNA containing telomeric sequences from Atlantic salmon (Salmo salar L.). At both low and high stringency conditions, a cloned probe containing (TTAGGG)n sequences binds only at the extreme termini of the chromosomes, not at interstitial sites. Blocks of interstitial telomere-like sequences were expected from the chromosome polymorphism because of Robertsonian translocations that exist in Atlantic salmon. Our results support the hypothesis that telomeric sequences were almost entirely lost during chromosome fusion events.
Similar content being viewed by others
Introduction
Telomeres are the physical ends of eukaryotic chromosomes that protect them from DNA degradation, end-to-end fusions, rearrangements and chromosome loss (Zakian, 1989; Blackburn, 1991; Smith & De Lange, 1997). All known telomeric sequences consist of simple repeated sequences of G- and C-rich complementary strands (Weiner, 1988). The G-rich DNA strand, orientated 5′–3′ towards the chromosome end, is synthesized by the telomerase enzyme, which is a specialized reverse transcriptase (Blackburn, 1991; Smith & De Lange, 1997). The human telomeric sequence (TTAGGG)n identified by Moyzis et al. (1988) shows a strong degree of evolutionary conservation with other vertebrates, as demonstrated in 91 different species in a systematic survey of hybridization sites of telomeric sequences in vertebrates (Meyne et al., 1989) showing that this sequence is the functional vertebrate telomere. The same telomeric repeat is present at the telomeres of trypanosomes (Van der Ploeg et al., 1984), slime moulds (Forney et al., 1987) and crustaceans (Pelliccia et al., 1994), probably constituting a universal telomeric consensus sequence.
The telomeric repeat sequence constitutes an excellent probe to study basic questions related to chromosomal evolution. According to their functions, telomeric sequences should be localized at the end of the chromosomes; nevertheless, nontelomeric blocks of the telomeric DNA sequences have been detected in many species of vertebrates from different taxonomic groups (Meyne et al., 1990) including avian species, Reptilia, Amphibia and Mammalia (marsupials, Insectivora, Chiroptera, Carnivora, Lagomorpha, rodents, Cetacea, Proboscidea, Artiodactyla and primates). The presence of telomeric DNA sequences at interstitial locations, very frequent in vertebrates, was explained by Meyne et al. (1990) as a consequence of short stretches of tandem telomeric sequences present in ancestral karyotypes as latent telomeres, or by fusion of chromosomes of ancestral species at the telomeres with subsequent amplification of retained stretches of telomeric sequence; in any case, rearrangement and amplification of this sequence within vertebrate genomes allows greater flexibility for karyotype changes by providing alternate sites for telomere formation.
Interstitial telomeric sequences are conserved in many vertebrates in which centric and/or tandem fusions appear to have occurred such as Indian muntjac (Muntiacus muntjak vaginalis) (Lee et al., 1993), chicken (Gallus domesticus) (Nanda & Schmid, 1994) and okapi (Okapia johnstoni). For this last species (with chromosome polymorphism 2n = 44–46 because of a recent Robertsonian fusion), Vermeesch et al. (1996) found a long interstitial telomeric element present at the fusion site of a fusion chromosome, interpreted as a remnant of the telomeres of ancestral chromosomes.
However, the conservation of telomeric sequences in centric fusion sites seems not to be a general phenomenon. Mouse chromosomes possessing multiple Robertsonian rearrangements were examined using fluorescent in situ hybridization (FISH) with the telomeric consensus sequence (Nanda et al., 1995); no hybridization signals were detected at the primary constriction of Robertsonian chromosomes; therefore the authors concluded that the formation of Robertsonian chromosomes in the mouse is associated with the loss of telomeric regions.
The salmonid karyotype most probably evolved from an ancestral tetraploidization event that occurred 50–100 million years ago (Ohno, 1970). Under this hypothesis, the chromosome number of the tetraploid ancestor was 96–100 acrocentric chromosomes. The different salmonid species then diverged as a consequence of diverse chromosome rearrangements and genome rediploidization, mainly involving centric fusions of nonhomologous chromosomes (Wright et al., 1983; Hartley, 1987; Pendas et al., 1994). Atlantic salmon (Salmo salar L.) has the lowest number of chromosome arms or fundamental number (NF = 74, 2n = 58). Wild populations present a conspicuous chromosome number polymorphism, that could be attributed to Robertsonian rearrangements (Hartley & Horne, 1984; Garcia-Vazquez et al., 1992). Studies on possible conservation of interstitial telomeric DNA sequences in this species will probably elucidate the basic mechanism of centric fusion, at least with respect to the extent of losses of telomeric DNA.
In this work we tested the presence of the hexanucleotide telomeric DNA sequences (TTAGGG)n in the genome of Atlantic salmon by means of cloning and sequencing, as well as its chromosomal location by means of FISH. In addition, information on the physical location of the cloned sequence was obtained by in situ hybridization using the whole cosmid vector that contains the telomeric sequence.
Materials and methods
Cosmid library
High molecular weight DNA was extracted from muscle tissue according to standard procedures (Taggart et al., 1992). This DNA was used for the construction of a total genomic library in a cosmid vector (sCos-1, Stratagene). Cosmids were isolated by plating out the library at low density and selecting after hybridization with the (TTAGGG) probe. The probe was labelled by a random priming technique using (α32P)dATP (Amersham). Filters were hybridized at 65°C in 3 × SSPE, 0.5% dried milk, 1% SDS and 6% polyethylene glycol overnight, and washed in 1 × SSC, 0.1% SDS at 65°C.
Cloning and nucleotide sequence
Positive cosmids were selected and digested with HaeIII and PstI; resulting fragments were electrophoresed in 2% agarose gels, transferred to nylon membranes and hybridized with the probe (TTAGGG)4. Positive bands were excised from the gel and DNA was eluted from the agarose using a silica matrix (Bio 101) and ligated in the SmaI site of the pUC18 cloning vector, then sequenced in both strands by using the Sequenase Kit (USB) with universal primers. The reaction products were fractionated by electrophoresis on a 6% polyacrylamide gel. Searches of the GenBank databases were carried out using the computer search and local similarity alignment program ‘FASTA’ (Pearson & Lipman, 1988).
Chromosome preparation and FISH
Blood was collected from the dorsal artery of 10 large fish, anaesthetized with MS-222, using a heparinized syringe. Metaphases were obtained from blood cultures as described by Sanchez et al. (1990).
Probes were labelled with biotin-11-dUTP using a nick translation kit (Boehringer). Slides were pretreated with RNAase and pepsin as described by Wiegant et al. (1991). Chromosomal DNA was denatured by immersing the slides at 80°C in 70% formamide 2 × SSC, pH 7.0 for 3 min. Slides were then dehydrated in an alcohol series (70%, 90%, 99% ethanol). The hybridization solution for high stringency conditions consisted of 50% formamide in 2 × SSC, 10% dextran sulphate and 500 μg/mL biotinylated DNA probe. For low stringency conditions formamide was reduced to 30%. After denaturation, 30 μL of hybridization mixture was applied to each slide under a sealed coverslip. Hybridization was performed in a moist chamber at 37°C overnight. Then slides were washed in 50% formamide at 45°C and 0.1 × SSC at 60°C (high stringency conditions) or 30% formamide at 45°C and 0.1 × SSC at 60°C (low stringency conditions). Hybridization was detected with fluorescein-conjugated avidin (Vector Laboratories). The signal was enhanced by incubation with biotinylated goat-antiavidin (Vector) followed by fluorescein-conjugated avidin. Chromosomes were counterstained with propidium iodide (1 μg/mL) and visualized and photographed in a Nikon Fluophot microscope on a 400 ASA Kodak colour slide film (high stringency hybridizations), and in a Zeiss axiophot fluorescent microscope equipped with a CCD camera (Photometrics) in the case of low stringency hybridizations.
In situ hybridization of the whole cosmid was carried out only under high stringency conditions to avoid background resulting from dispersed repeated sequences.
Results
Cloning of telomeric sequences
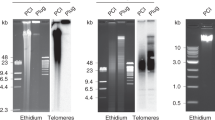
Positive clones were identified after screening the cosmid library with the (TTAGGG)4 probe. Five positive cosmids were selected, digested with HaeIII and PstI and hybridized again with the telomeric probe. Positive bands observed were longer than 5 kb in all cases. One of the positive bands (around 7 kb long) from the cosmid Tel-4 was cloned, digested with RsaI, AluI and DdeI, and probed with the telomeric probe. One positive band of 554 bp resulted from DdeI digestion. This band was subcloned in pUC18 and completely sequenced. The complete sequence (clone Tel-554) is shown in Fig. 1. Twelve quasi-perfect repeats of the (TTAGGG) motif were observed. Searches in the GenBank data files did not reveal any homology to the sequence adjacent to the telomeric repeat.
Chromosomal location of telomeric sequences
All Atlantic salmon individuals analysed showed 2n = 58 and NF = 74, the standard and fundamental chromosome number of Salmo salar (Phillips & Hartley, 1988).
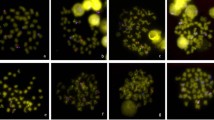
At least 20 cells per individual were evaluated for the presence of hybridization signal. In situ hybridization with the clone Tel-554 labelled all the Atlantic salmon chromosomes at their termini, both in high (Fig. 2a) and low (Fig. 2b) stringency conditions. Not all telomeres were clearly labelled in each metaphase; some telomeres showed only faint hybridization signal. Hybridization signals at interstitial chromosome locations were not observed in any case.
The whole cosmid Tel-4 was used for in situ hybridization to Atlantic salmon metaphase chromosomes (Fig. 3) under high stringency conditions to avoid background caused by dispersed repeated sequences. Hybridization signals appeared on almost all telomeric sites; some of them were bright and some were faint. In addition, large bright signals appeared on a whole arm of a medium-sized metacentric chromosome pair corresponding to the rDNA-bearing chromosome arm (Pendas et al., 1993).
Discussion
In this work we have demonstrated the presence of the telomeric DNA sequence (TTAGGG)n within the genome of Atlantic salmon by means of direct cloning and sequencing. Theoretically, cloning telomeres is complicated because fragments derived from the chromosome end have only one restriction enzyme-generated terminus, therefore they are rarely represented in a conventional DNA library (Zakian, 1989). Cloning telomeres is possible only in those species in which telomeric DNA sequences are also present at nontelomeric sites. In our case, the presence in the total genomic library of clones containing the (TTAGGG) telomeric repeat reveals the presence of this sequence at nontelomeric locations; the small number of repeats could correspond to subtelomeric amplifications of the telomeric repeats, as found by Vermeesch et al. (1996) for the giraffe, with short stretches or degenerate telomeric sequences present at intrachromosomal sites.
Alternatively, they could correspond to a small number of repeats conserved after centric fusions, as has occurred in some avian and mammalian species (Lee et al., 1993; Nanda & Schmid, 1994; Vermeesch et al., 1996), most probably in centromeric or interstitial positions.
Although different stringency conditions were used for FISH using the clone Tel-554, hybridization signals were visualized only at the chromosome ends. These results agree with Meyne et al. (1990), who found telomere-only patterns for several fishes (Salmoniformes and Cypriniformes). Gornung et al. (1998) also found hybridization signals of telomeric sequences restricted to telomeres in zebra fish (Danio rerio). From the existence of Robertsonian polymorphism in Atlantic salmon (Hartley & Horne, 1984; Garcia-Vazquez et al., 1992), the appearance of hybridization signals at centromeric or interstitial locations could be expected if telomeric sequences were conserved in fusion sites, as has occurred in the okapi (Okapia johnstoni), where interstitial telomeric DNA sequences are probably related to the recent origin of a chromosomal polymorphism (Vermeesch et al., 1996). Nevertheless, the case of Atlantic salmon seems to be different. It is possible that, as suggested by Nanda et al. (1995), substantial amounts of chromosomal DNA were eliminated during the formation of Robertsonian chromosomes, including telomeric sequences that could be almost entirely lost from the original chromosomes (Silva & Yonenage-Yassuda, 1997). It is possible that the number of repeats of telomeric DNA sequence located in nontelomeric regions in Atlantic salmon was so small that they produced extremely faint hybridization signals which would escape detection with the current resolution of FISH technology. Nevertheless, the methodology here employed to visualize hybridization signals at low stringency conditions (with a CCD camera) is powerful enough to detect small amounts of fluorescence; therefore problems derived from poor resolution are not expected.
Our results are contradictory to those found by Abuin et al. (1996), who detected hybridization signals of the telomeric sequence (TTAGGG)n at the telomeres of all chromosomes and at interstitial heterochromatic blocks in particular large chromosomes of Salmo salar under low stringency conditions (30% formamide). When they employed high stringency conditions they could not detect either telomeric or interstitial hybridization signals in any chromosome. The explanation for this contradiction could simply lie in the type of probe employed in FISH. In our case, we employed a true Atlantic salmon telomeric sequence obtained by direct cloning in a total genomic library, whereas Abuin et al. employed (TTAGGG)n oligonucleotides. They could have obtained heterologous hybridization with telomere-like sequences associated with heterochromatin or satellite DNA (in fact, our probe labelled chromosome ends also under high stringency conditions whereas their probe did not). Another possibility to explain the existence of large blocks of interstitial telomeric DNA sequences in some cases but not in others could be the existence of intraspecific differences in interstitial hybridization sites for Atlantic salmon, as demonstrated by Wiley et al. (1992) for hylid frogs. Absence of interstitial sites in some individuals and their presence in others in those amphibians was attributed to unequal crossing-over, submicroscopic deletion or differential amplification. A similar phenomenon could be also imagined in the present case.
The physical location of the cloned sequence, determined by in situ hybridizing the whole cosmid Tel-4, is the arm of the metacentric chromosome pair bearing rDNA genes (Pendas et al., 1993). In this arm, and as a part of the rDNA cistron, we have also found a large high-copy repeated DNA sequence (Moran et al., 1997). In other species, such as Atlantic eels (Anguilla anguilla and A. rostrata) (Salvadori et al., 1995), an association of telomere-like sequences and rDNA has been found. It is possible that the same situation applies to Atlantic salmon: some copies of telomeric sequences could have remained associated with rDNA. Alternatively, we can not exclude the possibility that Tel-4 contained, together with telomeric sequences, high-copy repeated sequences similar to those that map in the rDNA such as the BglI element (Moran et al., 1997).
In summary, we can conclude that the presence of the telomeric DNA sequence (TTAGGG) at telomeric regions has been demonstrated for Atlantic salmon. Such sequences are not present at nontelomeric regions at the resolution of in situ hybridization. Our results support the hypothesis of loss of telomeric sequences during centric fusions, the most probable type of rearrangement mechanism involved in Atlantic salmon chromosome evolution.
References
Abuin, M., Martinez, P. and Sanchez, L. (1996). Localization of the repetitive telomeric sequence (TTAGGG)n in four salmonid species. Genome, 39: 1035–1038.
Blackburn, H. (1991). Structure and function of telomeres. Nature, 350: 569–572.
Forney, J., Henderson, E. R. and Blackburn, E. H. (1987). Identification of the telomeric sequence of the acellular slime molds Didimium iridis and Physarum polycephalum. Nucl Acids Res, 15: 9143–9152.
Garcia-Vazquez, E., Moran, P., Pendas, A. M., Linde, A. R. and Izquierdo, J. I. (1992). Studies in individual spawnings of Salmo salar: a model to explain chromosome polymorphism patterns. Theor Appl Genet, 85: 61–67.
Gornung, E., Gabrielli, I. and Sola, L. (1998). Localization of the (TTAGGG)n telomeric sequence in zebrafish chromosomes. Genome, 41: 136–138.
Hartley, S. E. (1987). The chromosomes of the salmonid fishes. Biol Rev, 62: 197–214.
Hartley, S. E. and Horne, M. T. (1984). Chromosome polymorphism and constitutive heterochromatin in the Atlantic salmon, Salmo salar. Chromosoma, 89: 377–380.
Lee, C., Sasi, R. and Lin, C. C. (1993). Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs. Cytogenet Cell Genet, 63: 156–159.
Meyne, J., Ratcliff, R. L. and Moyzis, R. K. (1989). Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA, 86: 7049–7053.
Meyne, J., Baker, R. J., Hobart, H. H., Hsu, T. C., Ryder, O. A. and Ward, O. G. et al. (1990). Distribution of non-telomeric sites of (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma, 99: 3–10.
Moran, P., Reed, K. M., Perez, J., Oakley, T. H., Phillips, R. B., Garcia-Vazquez, E. and Pendas, A. M. (1997). Physical localization and characterization of the BglI element in the genomes of Atlantic salmon (Salmo salar L.) and brown trout (S. trutta L.). Gene, 194: 9–18.
Moyzis, R. K., Buckingham, J. M., Cram, L. S., Dani, M., Deaven, L. L. and Jones, M. D. et al. (1988). A highly conserved repetitive DNA sequence, (TTAGGG)n present at the telomeres of human chromosomes. Proc Natl Acad Sci USA, 85: 6622–6626.
Nanda, I. and Schmid, M. (1994). Localization of the telomeric (TTAGGG)n sequence in chicken (Gallus domesticus) chromosomes. Cytogenet Cell Genet, 65: 190–193.
Nanda, I., Schneider-Rasp, S., Winking, H. and Schmid, H. (1995). Loss of telomeric sites in the chromosomes of Mus musculus domesticus (Rodentia: Muridae) during Robertsonian rearrangements. Chromosome Res, 3: 399–409.
Ohno, S. (1970). The enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Trans Am Fish Soc, 99: 120–130.
Pearson, W. R. and Lipman, D. J. (1998). Improved tools for biological sequence comparison. Proc Natl Acad Sci USA, 85: 2444–2448.
Pelliccia, F., Volpi, E. V., Lanza, V., Gaddini, L., Baldini, A. and Rocchi, A. (1994). Telomeric sequences of Asellus aquaticus (Crust. Isop.). Heredity, 72: 78–80.
Pendas, A. M., Moran, P. and Garcia-Vazquez, E. (1993). Ribosomal RNA genes are interspersed throughout a heterochromatic chromosome arm in Atlantic salmon. Cytogenet Cell Genet, 63: 128–130.
Pendas, A. M., Moran, P. and Garcia-Vazquez, E. (1994). Organization and chromosomal location of the major histone cluster in brown trout, Atlantic salmon and rainbow trout. Chromosoma, 103: 147–152.
Philips, R. B. and Hartley, S. E. (1988). Fluorescent banding patterns of the chromosomes of the genus Salmo. Genome, 30: 193–197.
Salvadori, S., Deiana, A., Elisabetta, C., Floridia, G., Rossi, E. and Zuffardi, O. (1995). Colocalization of (TTAGGG)n telomeric sequences and ribosomal genes in Atlantic eels. Chromosome Res, 3: 54–58.
Sanchez, L., Martinez, P., Viňas, A. and Bouza, C. (1990). Analysis of the structure and variability of nucleolar organizer regions of Salmo trutta by C-, Ag- and restriction endonuclease banding. Cytogenet Cell Genet, 54: 6–9.
Silva, M. J., De, J. and Yonenaga-Yassuda, Y. (1997). New karyotypes of two related species of Oligoryzomys genus (Cricetidae, Rodentia) involving centric fusion with loss of NORs and distribution of telomeric (TTAGGG)n sequences. Hereditas, 127: 217–229.
Smith, S. and Delange, T. (1997). TRF1, a mammalian telomeric protein. Trends Genet, 13: 21–26.
Taggart, J. B., Hynes, R. A., Prodöhl, P. A. and Ferguson, A. (1992). A simplified protocol. for routine total DNA isolation from salmonid fishes. J Fish Biol, 40: 963–965.
Van der Ploeg, L. H. T., Lin, A. Y. C. and Borst, P. (1984). Structure of the growing telomeres of Trypanosomes. Cell, 36: 459–468.
Vermeesch, J. R. D. E., Meurichy, W., Van der Berghe, H., Marynen, P. and Petit, P. (1996). Differences in the distribution and nature of the interstitial telomeric (TTAGGG)n sequences in the chromosomes of the Giraffidae, okapi (Okapia johnstoni), and giraffe (Giraffa camelopardalis): evidence for ancestral telomeres at the okapi polymorphic rob (4;26) fusion site. Cytogenet Cell Genet, 72: 310–315.
Weiner, A. (1988). Eukaryotic nuclear telomeres: molecular fossils of the RNP world? Cell, 52: 155–157.
Wiegant, J., Ried, T., Nederlof, P. M., Vanderploeg, M., Tanke, H. J. and Raap, A. K. (1991). In situ hybridization with fluoresceinated DNA. Nucl Acids Res, 19: 3237–3241.
Wiley, J. E., Meyne, J., Little, M. L. and Stout, L. (1992). Interstitial hybridization sites of the (TTAGGG)n telomeric sequence on the chromosomes of some North American hylid frogs. Cytogenet Cell Genet, 61: 55–57.
Wright, E. J., Kenneth, J., Hollister, A. and May, B. (1983). Meiotic models to explain classical linkage, pseudolinkage and chromosome pairing in tetraploid derivative salmonid genomes. Genet Evol, 10: 239–260.
Zakian, A. (1989). Structure and function of telomeres. Ann Rev Genet, 23: 579–604.
Acknowledgements
We are indebted to Dr Alberto M. Pendas for kindly collaborating in this work and to Jose Luis Martinez for technical assistance. This work has been supported by the Spanish Grant DGICYT PB94–1327.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Perez, J., Moran, P. & Garcia-Vazquez, E. Cloning and physical mapping of Atlantic salmon (Salmo salar L.) telomeric sequences. Heredity 82, 409–414 (1999). https://doi.org/10.1038/sj.hdy.6884980
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884980
Keywords
This article is cited by
-
Cytogenetic investigation of Arctic char × brook trout F1, F2 and backcross hybrids revealed remnants of the chromosomal rearrangements
Journal of Applied Genetics (2021)
-
Conservation and characterization of unique porcine interstitial telomeric sequences
Science China Life Sciences (2012)
-
Independent fusions and recent origins of sex chromosomes in the evolution and diversification of glass knife fishes (Eigenmannia)
Heredity (2011)
-
Karyotypic characterization and genomic organization of the 5S rDNA in Erpetoichthys calabaricus (Osteichthyes, Polypteridae)
Genetica (2007)