Abstract
The genome size and the base composition (GC%) of eight Hypochaeris species were determined by flow cytometry in order to establish the pattern of nuclear DNA variation within the genus. The species analysed showed an almost fivefold range of variation from 1.68 pg in H. cretensis to 8.10 pg in H. uniflora. This variation in DNA content is greater between taxonomic sections of Hypochaeris species than within a section. There was no correlation between 2C DNA content and GC% indicating that neither the GC fraction nor the AT fraction were preferentially associated with variation in genome size. Because there is little heterochromatin, these results show that it is interspersed repeated sequences that are most probably implicated in this variation. From phylogenetic analysis, it is likely that genome size has evolved by loss of DNA content in some lineages and by gain in one lineage from an ancestral genome which was probably similar to genomes of intermediate size in Hypochaeris.
Similar content being viewed by others
Introduction
The genus Hypochaeris (Asteraceae) contains one species in Asia, about 10 species in Europe and the remaining 40 species in South America. This genus is considered by Stebbins et al. (1953) as having the most unusual geographical distribution of any in the tribe Lactuceae. Several hypotheses of karyotype and chromosome evolution within the genus have been suggested in earlier studies (Stebbins, 1971; Barghi et al., 1989; Siljak-Yakovlev et al., 1994; Cerbah et al., 1995, Ruas et al., 1995). European species display various chromosome numbers (2n=6, 8, 10, 12) and symmetrical or asymmetrical karyotypes, whereas South American species display a single basic chromosome number (x=4) and an asymmetrical bimodal karyotype. The karyological uniformity of South American species contrasts with a great diversity in their morphology and habitats.
Determination of nuclear DNA content is important in elucidating systematics and evolutionary relationships among species (Juan & Petitpierre, 1990; Srivastava & Lavania, 1991; Bharathan et al., 1994). The study of the genome size in closely related species can provide insight into the origin of DNA variation, but also into the direction of changes and the fraction of DNA involved in these changes (Bennett & Leitch, 1995). It is now established that the most important variation in DNA amount, apart from polyploidization and chromosome endoreplication, occurs in the repetitive fraction of the nuclear genome and that this variation can be under environmental selective pressure (Cullis, 1990). The genome can also rapidly change by the action of transposable elements which cause deletions or amplifications (Flavell, 1982). However, detailed knowledge about the mechanisms of change in DNA content and about factors determining the rates of amplification, deletions and rearrangements, remains lacking.
In the genus Hypochaeris, the variation observed in chromosome size (Mugnier & Siljak-Yakovlev, 1987; Barghi et al., 1989; Siljak-Yakovlev et al., 1994) suggests that there is significant variation in genome size. Detailed karyotypes of the species H. oligocephala, H.␣ achyrophorus and H. uniflora are here constructed for the first time from morphometric statistical data. 2C values of four South American species (H. chillensis (H.␣ B. K.) Hieron, H. microcephala (Sch. Bip.) Cabr. var. albiflora (O. K.) Cabr., H. megapotamica Cabr. and H.␣pampasica Cabr.) and two European species (H.␣ maculata L. and H. robertia Fiori) have been already published (Cerbah et al., 1995). It would be useful to know the variation within the whole genus. Hence, the genome size and base composition (GC%) of the studied species were determined by flow cytometry in order to establish the pattern of nuclear DNA variation among related species of Hypochaeris, as a contribution to the understanding of genome organization and evolution within this group.
Materials and methods
Eight species of Hypochaeris were analysed for DNA amount and GC%. Origins of samples are given in Table 1. Plants were grown in the experimental greenhouse of the University of Paris XI, Orsay campus. In this selection of species, all the basic chromosome numbers known for the genus were represented.
The Feulgen method was used to establish karyotypes of the species H. achyrophorus, H. oligocephala, and H. uniflora. For this purpose, chromosome arms were measured on 10 metaphase plates from different individuals: chromosome types, centromeric indices and arm ratios were determined according to Levan et al. (1964). Idiograms were drawn using the mean values. Asymmetry indices [AsI=(Σ long arms/total chromosome length) × 100] were calculated according to Arano & Saito (1980).
Total DNA amount in nuclei was assessed by flow cytometry using Petunia hybrida (2C=2.85 pg, 41% GC) as internal reference according to a technique adapted from Brown et al. (1991) and described in Cerbah et al. (1995). Total DNA content was determined by the ratio of the modal position of 2C stained nuclei of samples relative to those of Petunia hybrida cv. P×Pc6 (INRA, Dijon), or Pisum sativum cv. Long Express (Truffaut SA), using ethidium bromide (Sigma) at 30 μg mL−1. Two other fluorochromes, AT-specific bisbenzimide Hoechst 33342 (Aldrich Chimie) at 5 μg mL−1 and GC-specific chromomycin A3 (Serva) at 30 μg mL−1, were also used to compute base ratios. Five individuals per species were assessed for each analysis, which was made on about 5000 nuclei using an EPICS V cytofluorometer (Coultronics France, Margency). The average coefficient of variation of the 2C Hypochaeris nuclei in histograms was about 3% for ethidium bromide, 5% for Hoechst 33342 and 6.5% for chromomycin A3. Base ratios were calculated according the model of Godelle et al. (1993), which takes into account the number of fixation sites for fluorescence of each fluorochrome (on average a binding site for five A-T and three G-C base pairs, respectively, for Hoechst 33342 and chromomycin A3). Mean values of samples for total DNA amount and GC% were compared by ANOVA followed by a Tukey honest significant difference test for unequal n. 2C DNA content values for P. hybrida (2.85 pg) and P. sativum (8.37 pg) were taken from Marie & Brown (1993).
Results
Morphometric chromosome data for the species H.␣ uniflora, H. oligocephala and H. achyrophorus were used to construct idiograms (Fig. 1). The five chromosome pairs of H. uniflora were submetacentric, the two smallest pairs bearing a satellite on their short arm (Fig. 1a). According to the Asymmetry Index (67.7%) the karyotype of H. uniflora is symmetrical. Hypochaeris oligocephala had two metacentric chromosome pairs (1 and 2) and one subtelocentric pair (3) with a satellite on the short arm (Fig. 1b). This chromosomal region, including the satellite and its secondary constriction, is generally the chromosomal location of nuclear rDNA, also called the nucleolar organizer region (NOR). The karyotype of H. achyrophorus showed two submetacentric chromosomes pairs (1 and 6), two subtelocentric pairs (4 and 5) and two metacentric pairs (2 and 3). Chromosome pair 3 had a satellite on the long arm (Fig. 1c). This feature is reported here for the first time in the genus Hypochaeris, where species studied previously had satellites on the short arm of chromosomes.
Estimations of 2C DNA amounts and GC percentages are given in Fig. 2. Genome sizes of four South American species and two European species previously described (Cerbah et al., 1995) are included in the same figure for comparison. The species analysed showed an almost fivefold range of variation from 1.68 pg in H.␣ cretensis to 8.10 pg in H. uniflora. Comparison of results obtained by a Tukey-HSD-unequal n-test showed that differences in DNA amounts between all Hypochaeris species were highly significant (P<0.001) except for comparisons between H. maculata and H. illyrica and between H. robertia and H. radicata.
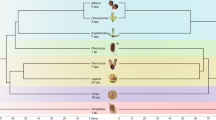
Phylogenetic ITS consensus tree of 14 Hypochaeris species with the two outgroups Leontodon hispidus and Hyoseris scabra from Cerbah et al. (1998b) with bootstrap values indicated above corresponding nodes. Morphological taxonomic section according to Hoffman (1891). aData published in Cerbah et al. (1995). bData published in Siljak-Yakovlev et al. (1994). cData published in Barghi et al. (1989). dData published in Mugnier & Siljak-Yakovlev (1987).
The GC% ranged from 37.9% to 44.1%, a 6% variation between species. There was no correlation between DNA content and GC%, although the 2C DNA content was significantly positively correlated (r=0.863, P<0.001) with the total chromosome length (TCL).
From the results, it was particularly striking that species with the highest chromosome number 2n=12 (H.␣achyrophorus and H. laevigata) did not have the highest DNA content. However, species which showed the smallest chromosome number 2n=6 (H.␣ cretensis and H. oligocephala) had among the smallest DNA contents.
Discussion
Karyological comparisons of H. uniflora and H. oligocephala with their respective close relatives do not reveal distinctive features. Hypochaeris oligocephala had a similar karyotype to H. cretensis, but a higher asymmetry index (AsI). Chromosomes of H. uniflora were of similar morphology to those of its two relatives H. maculata and H. illyrica but its AsI was lower and its TCL intermediate.
Hypochaeris achyrophorus showed more differences from its relative H. laevigata (2n=12). In H. achyrophorus, the TCL reflects the greater length of each chromosome pair and the chromosome pairs 4 and 5 were subtelocentric, increasing the AsI value (67.1%). This species showed only one chromosome pair bearing satellites (long arm of pair 3) instead of two pairs (short arms of pairs 2 and 3) in H. laevigata (Barghi et al., 1989). These differences could be attributed to chromosomal rearrangements involving the NORs during evolution.
In order to investigate evolutionary trends of genome size in these related species, a molecular phylogeny based on Internal Transcribed Spacer (ITS) sequences of nuclear ribosomal DNA (Cerbah et al., 1998b), was constructed. The phylogenetic consensus tree is shown in Fig. 2, where DNA contents, TLC, AsI, taxonomic sections and origin of species are added for comparison. Evolutionary trends previously proposed by Stebbins et al. (1953) for the genus could be summarized as follows: the original basic chromosome number of the genus Hypochaeris is x=5 and a decrease of the basic chromosome number is associated with an increase of karyotype asymmetry, as generally observed in the tribe Lactuceae. The ITS phylogeny suggests an alternative hypothesis of both decrease and increase of chromosome number during evolution of the genus Hypochaeris, from an ancestral basic chromosome number of x=4 (Cerbah et al., 1998b). In the same way, the present results of genome size showed that both decrease and increase of DNA content have occurred during evolution. Because there is little heterochromatin in the genome of Hypochaeris species (Cerbah et al., 1995, 1998a), we propose that genome size has evolved by variation in the amount of interspersed repeated sequences rather than by the amount of tandemly repeated sequences. This is not the same in other Asteraceae such as Crepis (Godelle et al., 1993) for which variation in the amount of heterochromatin between species is correlated with variation in DNA content.
Hypochaeris robertia, which had an intermediate value of 2C DNA (2.62 pg), has a position at the base of the phylogenetic tree (Fig. 2). We can consequently suppose that from a common ancestor with a genome size close to that of H. robertia, the genus has evolved by increasing the DNA amount in lineages of the South American and H. maculata groups, and decreasing the DNA amount in the lineage of H. cretensis.
Genome size variation in the genus Hypochaeris corresponded approximately to the grouping of species in morphological taxonomic sections. The genus was divided into five sections according to Hoffmann (1891). The section Achyrophorus, which includes all South American and some European species, showed the highest values of DNA amount in the genus (from 4.14 to 8.10 pg). The lowest values of 2C DNA have been found in the sections Metabasis (1.68 and 2.36 pg) and Seriola (2.34 pg). The sections Euhypochaeris, Seriola, and Robertia are represented by species having intermediate values of DNA content (from 2.62 to 3.13 pg). The variation of DNA content was 2.6-fold within species having a chromosome number of 2n=10, and was 1.8-fold within species with 2n=8 chromosomes. Lastly, the variation was greater among taxonomic sections than within a section, except for the section Achyrophorus, which shows two classes of values, one for South American species and the other for European species (Fig. 2), showing that geographical species divergence was accompanied by changes in genome size.
The species H. achyrophorus (2n=12) and H. laevigata (2n=12) were suggested to be tetraploid (Stebbins et al., 1953; Barghi et al., 1989). However, Brullo et al. (1977) consider these species as diploids with a basic chromosome number of x=6, which is supported by the molecular phylogeny (Cerbah et al., 1998b). Moreover, H. oligocephala (2n=6; 2.36 pg) and H. laevigata (2n=12; 2.34 pg) had very similar DNA contents. Nevertheless, because H. achyrophorus has a DNA content of about 1.6-fold that of H. cretensis (2n=6), it could be that this species is derived by allotetraploidy. A loss of DNA following polyploid formation has been demonstrated in many studies (Raina et al., 1994; Feldman et al., 1997) but more experiments are needed to determine if this has occurred in Hypochaeris.
There was no overall significant correlation between 2C DNA content and GC%, indicating that neither the GC fraction nor the AT fraction was preferentially associated with variation of genome size. However, the variation of DNA content between H. cretensis and H.␣ oligocephala on the one hand, and H. achyrophorus and H. laevigata on the other, is associated with an increase in the GC fraction. In contrast, variation between H.␣ glabra and H. radicata is associated with an increase in the AT fraction.
Hypochaeris cretensis and H. oligocephala, which showed the smallest DNA content, are very closely related to the group of H. maculata, which had the largest genome (Fig. 2. This implies that rapid size variation can arise in related species, as noticed by Cullis (1990).
Selection pressures such as environmental factors could have increased or decreased the DNA amounts. A decrease of DNA content, in the case of H. cretensis and H. oligocephala, associated with a specialization for a particular habitat, could have taken place during evolution. The natural ecological site for H. oligocephala is on cliffs in Tenerife (Lack, 1978) and for H. cretensis, on rocks (Brullo et al., 1977). On the other hand, in the H. maculata group an increase of DNA content has occurred which could be associated with an adaptation to higher altitudes, as has been shown for Zea mays ssp. mays (Rayburn & Auger, 1990) and for the Crepis praemorsa complex of species (Godelle et al., 1993). Hypochaeris illyrica and H. uniflora have significantly more DNA than H. maculata and are found at higher altitudes in mountain habitats.
Following these results, analysis of repeated interspersed DNA sequence families should provide a better understanding of genome evolution in Hypochaeris species.
References
Arano, H., Saito, (1980). Cytological studies in family Umbelliferae 5. Karyotypes of seven species in subtribe Seselinae. Kromosomo II, 17: 471–480.
Barghi, N., Mugnier, C. and Siljak-Yakovlev, S. (1989). Karyological studies in some Hypochaeris spp. (Compositae) from Sicily. Pl Syst Evol, 168: 49–57.
Bennett, M. D. and Leitch, I. J. (1995). Nuclear DNA amounts in Angiosperms. Ann Bot, 76: 113–176.
Bharathan, G., Lambert, G. and Galbraith, D. W. (1994). Nuclear DNA content of monocotyledons and related taxa. Am J Bot, 81: 381–386.
Brown, S. C., Bergounioux, C., Tallet, S. and Marie, D. (1991). Flow cytometry of nuclei for ploidy and cell cycle analysis. In: Negrutiu, I. and Gharti-Chherti, G. B. (eds) A Laboratory Guide for Cellular and Molecular Plant Biology, pp. 326–345. Birkhaüser, Basel.
Brullo, S., Majorana, G., Pavone, P. and Terrasi, M. C. (1977). Numeri cromosomici per la Flora Italiana. Inf Bot Ital, 9: 283–298.
Cerbah, M., Coulaud, J., Godelle, B. and Siljak-Yakovlev, S. (1995). Genome size, fluorochrome banding, and karyotype evolution in some Hypochaeris species. Genome, 38: 689–695.
Cerbah, M., Coulaud, J. and Siljak-Yakovlev, S. (1998a). rDNA organization and species relationships in the genus Hypochaeris. J Hered, 89: 312–318.
Cerbah, M., Souza-Chies, T., Jubier, M. F., Lejeune, B. and Siljak-Yakovlev, S. (1998b). Molecular phylogeny of the genus Hypochaeris using internal transcribed spacers of nuclear rDNA: inference for chromosomal evolution. Mol Biol Evol, 15: 345–354.
Cullis, C. A. (1990). DNA rearrangements in response to environmental stress. Adv Genet, 28: 73–97.
Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A. A. and Vega, J. M. (1997). Rapid elimination of low-copy sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics, 147: 1381–1387.
Flavell, R. B. (1982). Sequence amplification, deletion and rearrangement: major sources of variation during species divergence. In: Dover, G. A. and Flavell, R. B. (eds) Genome Evolution, pp. 301–323. Academic Press, London.
Godelle, B., Cartier, D., Marie, D., Brown, S. C. and Siljak-Yakovlev, S. (1993). Heterochromatin study demonstrating the non-linearity of fluorometry useful for calculating genomic base composition. Cytometry, 14: 618–626.
Hoffmann, O. (1891). Liguliflorae-Cichorieae. In: Engler, A. and Prantl, K. (eds) Die Natürliche Pflanzenfamilien, IV, 5, pp. 350–387. Leipzig.
Juan, C. and Petitpierre, E. (1990). Evolution of genome size in darkling beetles (Tenebrionidae, Coleoptera). Genome, 34: 169–173.
Lack, H. W. (1978). Die Gattung Heywoodiella Svent. & Bramw. (Asteraceae, Lactuceae). Willdenowia, 8: 329–339.
Levan, A., Fredga, K. and Sandberg, A. A. (1964). Nomenclature for centromeric position on chromosomes. Hereditas, 52: 201–220.
Marie, D. and Brown, S. C. (1993). A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol Cell, 78: 41–51.
Mugnier, C. and Siljak-Yakovlev, S. (1987). Karyological study in some Yugoslavian populations of Hypochoeris (Compositae). Caryologia, 40: 319–324.
Raina, S. N., Parida, A., Koul, K. K., Salimath, S. S., Bisht, M. S., Raja, V. and Khoshoo, T. N. (1994). Associated chromosomal DNA changes in polyploids. Genome, 37: 560–564.
Rayburn, A. L. and Auger, J. A. (1990). Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theor Appl Genet, 79: 470–474.
Ruas, C. F., Ruas, P. M., Matzenbacher, N. I., Ross, G., Bernini, C. and Vanzela, A. L. L. (1995). Cytogenetic studies of some Hypochoeris species (Compositae) from Brazil. Am J Bot, 82: 369–375.
Siljak-Yakovlev, S., Bartoli, A., Roitman, G., Barghi, N. and Mugnier, C. (1994). Etude caryologique de trois espèces d'Hypochaeris originaires d'Argentine: H. chiliensis (H. B. K.) Hieron, H. megapotamica Cabr. et H. microcephala (Sch. Bip.) Cabr. var. albiflora (O.K.) Cabr. Can J Bot, 72: 1496–1502.
Srivastava, S. and Lavania, U. C. (1991). Evolutionary DNA variation in Papaver. Genome, 34: 763–768.
Stebbins, G. L. (1971). Chromosomal Evolution in Higher Plants, Edward Arnold, London.
Stebbins, G. L., Jenkins, J. A. and Walters, M. S. (1953). Chromosomes and phylogeny in the Compositae, tribe Cichorieae. Univ. Calif. Publ Bot, 26: 401–430.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cerbah, M., Coulaud, J., Brown, S. et al. Evolutionary DNA variation in the genus Hypochaeris. Heredity 82, 261–266 (1999). https://doi.org/10.1038/sj.hdy.6884690
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884690
Keywords
This article is cited by
-
Genome size and chromosome number of ten plant species from Kerguelen Islands
Polar Biology (2020)
-
Molecular–cytogenetic studies of ribosomal RNA genes and heterochromatin in three European Fraxinus species
Tree Genetics & Genomes (2014)
-
Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa
Plant and Soil (2013)
-
Genome size in Filago L. (Asteraceae, Gnaphalieae) and related genera: phylogenetic, evolutionary and ecological implications
Plant Systematics and Evolution (2013)
-
Genome size and cytogenetic characterization of three Algerian Retama species
Tree Genetics & Genomes (2011)