Abstract
Compatible data from 11 polymorphic allozyme loci were assembled from 14 published studies and additional unpublished information for 232 native collections of brown trout sampled over a broad geographical range. Most of the genetic variation was explained by the distribution of allelic variation at the LDH-C* and CK-A1* loci. Patterns of geographical distribution coupled with postglacial geological events supported a model of colonization from three preglacial lineages, each fixed for different sets of alleles at these two loci. During glacial retreat, recolonization is proposed to have occurred mainly into adjacent areas through (i) a north-western migration from an eastern Mediterranean–Caspian refuge, (ii) a northern expansion from a refuge in Atlantic drainages of Iberia and southern France, and (iii) a northern and eastern radiation from a refuge centred near the English Channel. Extant populations in deglaciated areas are suggested to represent mixed or pure descendants of these migrating groups. Repopulation from a fourth Mediterranean refuge distinguished by the presence of the LDH-A2*100QL allele was excluded based on the absence of this allele in repopulated areas.
Similar content being viewed by others
Introduction
The brown trout Salmo trutta L. is native to Eurasia and North Africa. The species is considered to be a component of the northern ichthyofauna, but populations from nonglaciated areas have persisted much longer than those occupying recently repopulated northern areas following glacial retreat (Ferguson, 1989). These refugial populations served as sources for recolonization since the end of the last glacial period about 13 000 years BP. However, an understanding of the differences among refuges and their patterns of postglacial migrations has emerged only during the past decade through the accumulation of biochemical and molecular genetic information. An initial two-population model of recolonization (Ferguson & Fleming, 1983; Hamilton et al., 1989) is now considered to be more complex (Hynes et al., 1996).
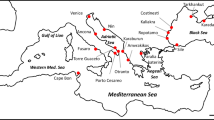
This paper seeks further clarification of glacial refugia and the process of postglacial colonization. We begin with current understanding arising from genetic data indicating the existence of multiple major lineages among the European brown trout populations (Fig. 1). Two lineages (I and II) among Atlantic drainages are characterized, respectively, by high allelic frequencies of LDH-C*90 and of CK-A1*115 (Hamilton et al., 1989; García-Marín & Pla, 1996). MtDNA haplotypes distinguish populations of lineage III in the Black, Caspian and Aral seas and those of the more western lineage IV in the Mediterranean Sea (Osinov & Bernatchez, 1996). The mtDNA data further distinguish other groupings of brown trout within lineage IV (Bernatchez et al., 1992; Apostolidis et al., 1997).
Distribution of presumed major lineages (I, II, III and IV) of brown trout during the latest glacial maximum (about 13000 years BP, indicated by dashed line) based on published allozyme and mtDNA data. Arrows identify proposed directions of migration during postglacial recolonization from lineages I, II and III.
Many questions remain unanswered about the boundaries and phylogeny of these groupings and of possible further major divisions of brown trout. This uncertainty extends to understanding the relative contributions of these lineages to the recolonization of deglaciated areas. We have assembled a set of compatible genetic data over a large part of the natural range of brown trout including unglaciated and recolonized areas. We use data of 11 polymorphic allozyme loci for 232 European brown trout samples obtained from several published studies, as well as unpublished data from our laboratory. From the analysis of these combined data, we consider the dynamics of postglacial colonization by brown trout.
Materials and methods
Selection of populations
Considerable allozyme data on populations of brown trout within the species' native range have accumulated during the past 20 years. In assembling the present data set, we had to use not more than 250 samples (the limitation of the BIOSYS-1 program used) from more than 500 reported samples. In selecting data, we focused on the following criteria.
Native populations
Samples were excluded where authors suspected introgression with or displacement by exogenous fish to be likely (but see Table 4). Similarly, hatchery populations were generally excluded except for the Gauya and Kura stocks (Osinov, 1984), which represented the first generation of local native spawners.
Reliability and extent of data
Studies were excluded where only a few loci had been examined, or where we questioned the validity of allelic or locus nomenclature to be compared.
Geographical range
Surveys covering a broad geographical range, or localities not previously examined, were given preference over those only examining areas included in other studies.
Based on these criteria, we used data representing presumably native fish from 232 discrete locations: 90 from the British Isles (Fleming, 1983; Crozier & Ferguson, 1986; Ferguson & Taggart, 1991; Cross et al., 1992); 15 from France (Krieg & Guyomard, 1985; Barbat-Leterrier et al., 1989); seven from Greece (Karakousis & Triantaphyllidis, 1990); 51 from Norway (Skaala & Jørdstad, 1987; Skaala & Nëvdal, 1989; Hindar et al., 1991; Skaala, 1992); six from Russia (Osinov, 1984); 33 from Sweden (Ryman, 1983); and 30 from Spain (García-Marín & Pla, 1996; and 6 unpublished locations). A detailed description of these collections, including the allele frequency for the most widespread variant at the four more polymorphic loci is available from the authors on request or accessible in the URL entry: http://fc.udg.es/~genetica/LIG/curri/appendix/tabla1.html. Only 46 out of the 232 samples were from the southern range, including six from France, three from Russia, and all of the Greek and Spanish collections; this disproportionality reflects a more recent and less intensive focus on southern populations.
Standardization and selection of loci and alleles
We initially considered 45 loci reported in the reviewed literature to be polymorphic. Most of these loci were excluded from the final data set because they were not examined in all studies. Others (e.g. esterases, malic enzyme) were excluded because of the difficulty in establishing homologies of loci and alleles among the different published sources. Ultimately, we chose a set of 11 loci (Table 1) in which we had confidence in the compatibility of data reported for the 232 selected locations: CK-A1*, G3PDH-2*, GPI-A*, GPI-B2*, IDHP-2*, IDHP-3*, LDH-A1*, LDH-A2*, LDH-C*, sMDH-A2* and sMDH-B1,2*.
Following the practice in brown trout genetic studies, all variation at the sMDH-B1,2* isolocus was assigned to sMDH-B1*; variation assigned simultaneously to both isoloci is restricted to a few Spanish rivers (García-Marín & Pla, 1996). Thus the sMDH-B2* locus was excluded from the present data set. Ferguson & Taggart (1991) observed variation in some collections at the sMDH-B1,2* but these authors did not give the allelic data, so fixation of the *100 alleles was assumed in our computations. A similar assumption was made with the cited but unreported variation at the IDHP-3* locus in Skaala (1992).
Data analysis
Components of gene diversities (HT, FST) for individual and all loci (Wright, 1978) were calculated for samples within each of the seven regions (British Isles, France, Greece, Norway, Russia, Sweden, Spain) and for all 232 samples (Tables 2 and Table 3). As an Euclidean measure (Sneath & Sokal, 1973), a pairwise matrix of genetic identities (I; Nei, 1978) was used for principal coordinates analysis (PCO; Gower, 1966). The likelihood of introgression among major ancestral groups during recolonization (e.g. Hamilton et al., 1989; Hynes et al., 1996) where geographical variation may take nonhierarchical forms, prompted the use of an ordination technique (PCO) rather than hierarchical cluster analysis (Lessa, 1990; Sokal, 1991).
Results
Allelic distribution of primary informative loci
The degree of genetic differentiation over the geographical range of sampled locations varied considerably among the 11 selected polymorphic loci. Means and indices of differentiation (FST) of allele frequencies varied among loci (Table 2). Four loci, CK-A1*, G3PDH-2*, LDH-C* and sMDH-A2*, had mean frequencies less than 0.90 for the *100 allele, and were therefore considered the most variable of the 11 selected loci. Allele frequencies at these loci varied considerably among the seven regions, with fixation of alternate alleles occurring in one or more populations at three of them (Table 3). Geographical patterns were particularly evident for LDH-C* and CK-A1*, which among the 11 loci had the largest variance among locations. The samples from France were particularly diverse at LDH-C*, having an FST of 0.880, with four of 15 locations fixed for the *90 allele, and six for the *100 allele. The LDH-C*100 allele predominated in the southern European localities, although intermediate frequencies in the Russian samples from the White Sea are notable exceptions. The LDH-C*90 allele prevailed in the British Isles, being fixed in 53 of 90 samples, and having a minimum allele frequency (0.350) larger than frequencies in Greek and Spanish collections.
Diversity of the CK-A1* locus was highest in Spain, France and Russia. Only the *100 allele was observed in the seven Greek samples. Fixation of the *115 allele in eight Spanish and one French sample was consistent with this allele serving as a marker for a south-western glacial refuge for brown trout (García-Marín & Pla, 1996). Also noteworthy is the high frequency of the *115 allele in some Swedish, Norwegian and Russian samples (0.609–0.916), in contrast to a maximum value of 0.500 in the British Isles.
Plots of principal coordinates based on the genetic identity matrix for the 11 polymorphic loci (Fig. 2) complement the data of Table 3. Values of PCO-1 explained 37% of the variability of the matrix and were strongly influenced by the LDH-C* locus. PCO-1 was positive for collections in areas of the lineages II, III and IV (Fig. 1) and for postglacial repopulated populations having high frequencies of LDH-C*100. Conversely, negative values located lineage I and postglacial populations where LDH-C*90 predominated. PCO-2 explained 17% of the variability of the matrix and roughly corresponded to frequencies for CK-A1*. High CK-A1*115 frequencies in lineage II distinguish these populations from those of adjacent lineages III and IV, which are typified by high frequencies of the CK-A1*100 allele. Similarly, PCO-2 separated populations with high CK-A1*115 allele frequency in areas of glacial retreat.
Discussion
Generalities arising from the examination of these compatible data compiled from many studies include the following points. 1 At the regional level, major features of genetic diversity of native populations of brown trout sampled throughout much of its range are reflected in the allelic distributions of LDH-C* and CK-A1* among the 11 polymorphic loci examined. The Discussion will focus on the allelic distributions of LDH-C* and CK-A1*, given the major influence of these two loci on the topography of relationships shown in Fig. 2 and on the allelic diversity among locations as presented in Table 3.
2 The allelic distributions appear to define three major geographical groups in unglaciated areas: (i) a north-western Atlantic group characterized by high frequencies of LDH-C*90 particularly in the British Isles; (ii) a Mediterranean group characterized by LDH-C*100 and CK-A1*100 alleles; and (iii) a south-western Atlantic group characterized by LDH-C*100 and CK-A1*115 alleles.3 Alleles distinguishing unglaciated regions occur at diverse localities and with varying frequencies throughout the repopulated northern range.
Additional studies
The above observations can be augmented by considering similar studies of Danish (Hansen et al., 1993), German (Riffel et al., 1995) and Swiss (Largiardèr & Scholl, 1995, 1996) populations that have been excluded so far because of more than a century of hatchery stocking and translocations involving exogenous fish. As summarized in Table 4, the allelic distributions in these 48 additional locations are typical of those from unstocked areas selected for this study, but with evidence of varying degrees of interbreeding with hatchery populations of German (presumably Rhenish) origin (García-Marín et al., 1991). Particularly noteworthy is the lower overall frequency of the LDH-C*90 allele in the Black Sea (Danube) and Mediterranean rivers, drainages where this allele is absent or at a low frequency in populations selected for this study. Also pertinent is the exclusive presence of the LDH-A2*100QL allele in a tributary of the upper Rhone river (data in Largiadèr & Scholl, 1996), as discussed in greater detail below.
Glacial refugia
The presumed lineages I–IV (Fig. 1) require consideration from the perspective of the present data. Concerning lineages I and II, we envisage two initially isolated groups characterized by fixation of different sets of alleles at LDH-C* and CK-A1*. Although polymorphism for the diagnostic alleles is possible within each putative refuge, allelic fixation appears to provide reliable tags in colonizing fish, given the regional generality of these distributions, and the tendency of brown trout populations to approach fixation relative to other salmonids (e.g. Ryman, 1983). Considering the present distribution of locations with higher LDH-C*90 and CK-A1*100 allele frequencies, a glacial refuge for lineage I is proposed in unglaciated areas of the southern British and adjacent continental drainages (e.g. Rhine, Meuse, Thames and Scheldt rivers). This refuge is consistent with the palaeohistory of the north-western European rivers during the most recent glacial maximum and low eustatic sea level, when a confluence of these rivers flowed through the Dover Strait into the English Channel (Gibbard, 1988).
To the south, a refuge for lineage II is proposed in the Atlantic drainages of the western Iberian Peninsula. Populations of this refuge are characterized by the fixation of the LDH-C*100 and CK-A1*115 alleles. Less glaciation in this area permitted the perpetuation of trout populations even during glacial peaks. Allelic distributions in Atlantic drainages of France suggest a glacial or early postglacial interaction of the northern (I) and western Iberian (II) lineages. An extension of lineage II to include French rivers entering the Bay of Biscay is supported by high LDH-C*100 and CK-A1*115 frequencies in this region (Kernec, Scorff and Elorn streams; Krieg & Guyomard, 1985). In more northern French drainages, intermediate CK-A1*115 frequencies suggest admixture of lineages I and II.
Mediterranean rivers are proposed as the refuge for lineage IV, and Black Sea and Caspian drainages for lineage III (Fig. 1). The fixation of the CK-A1*100 allele distinguishes these two refuges from lineage II populations. The restricted occurrence of the LDH-A2*100QL allele in the Mediterranean basin (Apostolidis et al., 1996; García-Marín & Pla, 1996; Largiardèr & Scholl, 1996) provides a useful marker to distinguish populations of lineage IV from those of lineage III. The absence of this allele from postglacially colonized regions and the palaeohydrographical features associated with the geography of this fourth lineage suggest little if any contribution to the colonization of northern deglaciated areas.
Proposed dynamics of recolonization
Mechanism of colonization
The following discussion is guided by the above approximations whereby three major glacial refuges (I, II, III) are characterized by different combinations of fixed genotypes at the loci LDH-C* and CK-A1*. Given these qualifications, the allelic distributions of these two loci in postglacially repopulated areas provide useful clues to the populations' ancestries (Table 5).
We propose a pattern of colonization based on a rapid, simultaneous spreading of brown trout from the glacial refugia, as suggested for postglacial range expansions in a variety of organisms (Hewitt, 1996). The genetic consequences expected when new colonies are founded by individuals from nearby populations modelled by Ibrahim et al. (1996) demonstrate that rare long-distance migration during range expansions leads to the establishment of pocket populations well in advance of the main wave that may persist for hundreds of generations. In brown trout, occasional long-distance migrants from lineages I, II or III would be able to establish colonies well ahead of the main distribution. This form of spreading may involve a series of population bottlenecks and may lead to homozygosity in the colonizing genomes (Nei et al., 1975). The persistence of such pioneer populations would be favoured by postglacial isostatic uplift that may have isolated individuals in upstream regions before the main colonizing wave arrived. Lineage I individuals, being nearest to the deglaciated areas, would have provided the main wave of colonization.
Eastern drainages During the preceding period of maximal glaciation (18000 years BP) many north-flowing rivers of eastern Europe and western Siberia were impounded by the Würmian ice-sheet. The ice-dammed lakes and ice-marginal streamways discharged southward into the Aral, Caspian and Black seas and then continued further into the Mediterranean (Grosswald, 1980; Gibbard, 1988). About 13500 years BP this radial drainage system was transformed into a single marginal one that extended westwards into the Norwegian Sea. These events permitted northward migration of brown trout of lineage III and their introduction into northern Europe early in the postglacial. The presence of LDH-C*100 at intermediate and high frequencies in the White Sea and some Norwegian samples presumably reflects this initial eastern invasion. From these bridgehead populations, lineage III expanded to other deglaciated areas as indicated by the presence of British populations with fixed or high frequency for the LDH-C*100 and CK-A1*100 alleles.
Atlantic drainages As glacial retreat proceeded from about 13 500 years BP, the most likely source of recolonization was the nearest refuge (Hewitt, 1996). The high frequency of LDH-C*90 in the British Isles clearly indicates colonization from unglaciated southern Britain (lineage I). The polymorphism at the LDH-C* locus in 37 out of 90 British samples and the presence of CK-A1*115 at low to moderate frequencies in 36 of these samples presumably reflects additional influence from lineage II. The absence of this latter allele in areas with significant presence of LDH-C*100 suggests additional influence from lineage III in the British Isles as indicated for eastern drainages. The existence of sympatric populations in the British Isles (e.g. Ferguson & Taggart, 1991) is proposed to reflect early colonizations and interactions from the three sources with subsequent divergences. As noted above, moderate CK-A1*115 and high LDH-C*90 frequencies in populations of northern France suggest possible admixture of lineages I and II in this region during the glacial period.
Norwegian populations in Atlantic drainages predominantly have high LDH-C*90 and low CK-A1*115 frequencies (Table 3). This pattern is particularly evident among anadromous populations having a greater opportunity for contemporary gene flow. As in the British Isles, major colonizations by individuals originating from the closer lineage I is presumed. Pure lineage I ancestry is suggested for populations fixed for LDH-C*90 and CK-A1*100 (e.g. Lake Vatnastølsvatn; Skaala & Nëvdal, 1989), whereas admixture of lineages I and II presumably has occurred in many of the remaining samples. The greater allele frequency variation at both marker loci among resident populations is attributed to higher levels of genetic drift expected in smaller populations.
Baltic drainages Until 10 000 years BP, the Baltic sea was an ice-marginal lake (Baltic Ice-lake) with no connection to the North Sea. Presumably, this lake was colonized initially by a lineage III invasion. After this time, the Baltic basin became a bay of the Atlantic with a connection through the present Lake Vänern. The open marine connection permitted an influx of several marine fishes, including, we propose, brown trout from lineages I and II during the colonization of other recently deglaciated areas of western Europe proposed by Hamilton et al. (1989). Beginning about 7500 years BP, isostatic uplift isolated many of the newly founded Baltic populations above waterfalls, and the Baltic basin itself gradually evolved towards its presently more isolated connection with the Atlantic ocean.
The populations of Norway, Sweden and Russia in Baltic drainages thus presumably reflect potential ancestries from lineages I, II and III. The genetic heterogeneity of Baltic populations supports such a diverse ancestry (Fig. 2. The resident Swedish populations indicate a generally high lineage I ancestry, with varying influence of lineage II based on the putative marker alleles of the two lineages. Mixed ancestries of lineages I and III are inferred for the Pleutajokk and Slorudsålvan samples, and of lineages II and III for the Sipmegejaure sample (Ryman, 1983).
Contrast with other models of recolonization
Details of recolonization distinguish the present model from others that have been based on more restricted sampling and, in some instances, on different procedures. Our model agrees with that of Hamilton et al. (1989) in identifying northern and southern sources of recolonization. However, instead of a late-arriving and vaguely defined ‘modern’ northern race that displaced an ‘ancestral’ southern one in recolonized regions, we assume the coexistence of multiple brown trout lineages before the last glaciation, with at least three lineages spreading simultaneously to NW Europe after deglaciation. Our model agrees with the mtDNA sequence data suggesting multiple lineages (at least five) for the population structure of brown trout in Europe, these mitochondrial data also suggesting a time of divergence of such lineages of 500000 years BP or older (Bernatchez et al., 1992; Apostolidis et al., 1997).
As stated above, our model that initial colonization preferentially occurred from the most proximal refuge(s), and that extant populations reflect the genotypes of these refugia, has been suggested for other organisms (Hewitt, 1996). Both glacial contractions and postglacial range expansion may produce homozygosity. Thus the fixation of LDH-C*90 in our proposed lineage I does not require a selective advantage for this allele during or prior to the colonization process, although it does not exclude this possibility.
Our hypothesis of an early northward migration of lineage III populations into the White Sea and the Baltic basin was also suggested by Bernatchez & Osinov (1995) based on mtDNA and allozyme data for populations in these regions. Similarly, our model supports the conclusion of Hynes et al. (1996) based on mtDNA analyses of samples collected over a broad geographical range that ‘postglacial colonization of brown trout in NW Europe was more complex than the dual colonization model which has been proposed on the basis of differential LDH-C* allele distribution’.
Certainly, clarification of the details of postglacial recolonization of brown trout awaits collection and analysis of additional information. Also, a model based on two loci with different distributions of fixed alleles in three refuges, plus a third locus that excludes colonization by a fourth lineage, masks large amounts of highly relevant information. Nevertheless, we feel that this model provides a useful alternative hypothesis for future investigations.
References
Apostolidis, A., Karakousis, Y. and Triantaphyllidis, C. (1996). Genetic divergence and phylogenetic relationships among Salmo trutta L. (brown trout) populations from Greece and other European countries. Heredity, 76: 551–560.
Apostolidis, A. P., Triantaphyllidis, C., Kouvatsi, A. and Economidis, S. (1997). Mitochondrial DNA sequence variation and phylogeography among Salmo trutta L. (Greek brown trout) populations. Mol Ecol, 6: 531–542.
Barbat-Leterrier, A., Guyomard, R. and Krieg, F. (1989). Introgression between introduced domesticated strains and Mediterranean native populations of brown trout (Salmo trutta L.). Aquat Living Resour, 2: 215–223.
Bernatchez, L. and Osinov, A. (1995). Genetic diversity of trout (genus Salmo) from its most eastern native range based on mitochondrial DNA and nuclear gene variation. Mol Ecol, 4: 285–297.
Bernatchez, L., Guyomard, R. and Bonhomme, F. (1992). DNA sequence variation of the mitochondrial control region among geographically and morphologically remote European brown trout Salmo trutta populations. Mol Ecol, 1: 161–173.
Cross, T. F., Mills, C. P. R. and Decourcy Williams, M. (1992). An intensive study of allozyme variation in freshwater resident and anadromous trout Salmo trutta L., in Western Ireland. J Fish Biol, 40: 25–32.
Crozier, W. W. and Ferguson, A. (1986). Electrophoretic examination of the population structure of brown trout Salmo trutta L., from the Lough Neagh catchment, Northern Ireland. J Fish Biol, 28: 459–477.
Ferguson, A. (1989). Genetic differences among brown trout Salmo trutta stocks and their importance for the conservation and management of the species. Freshwater Biol, 21: 35–46.
Ferguson, A. and Fleming, C. C. (1983). Evolutionary and taxonomic significance of protein variation in the brown trout (Salmo trutta L.). In: Oxford, G. S. and Rollinson, D. (eds) Protein Polymorphism: Adaptive and Taxonomic Significance, special vol. 24, Systematics Association, pp. 85–99. Academic Press, London.
Ferguson, A. and Taggart, J. B. (1991). Genetic differentiation among sympatric brown trout (Salmo trutta) populations of Lough Melvin, Ireland. Biol J Linn Soc, 43: 221–237.
Fleming, C. C. (1983). Population Biology of Anadromous Brown Trout (Salmo trutta L.) in Ireland and Britain. Ph.D. Thesis, Department of Zoology, the Queen's University of Belfast.
Garcáa-Marín, J. L. and Pla, C. (1996). Origins and relationships of native populations of brown trout (Salmo trutta) in Spain. Heredity, 77: 313–323.
García-Marín, J. L., Jorde, P. E., Ryman, N., Utter, F. and Andpla, C. (1991). Management implications of genetic differentiation between native and hatchery populations of brown trout (Salmo trutta) in Spain. Aquaculture, 95: 235–249.
Gibbard, P. L. (1988). The history of the great Northwest European rivers during the past three million years. Phil Trans R Soc B, 318: 554–602.
Gower, J. C. (1966). Some distance properties of latent root and vector methods in multivariate analysis. Biometrika, 53: 315–328.
Grosswald, M. G. (1980). Late Weichselian Ice sheet of Northern Eurasia. Quaternary Res, 13: 1–32.
Hamilton, K. E., Ferguson, A., Taggart, J. B., Tómason, T., Walker, A. and Fahy, E. (1989). Post-glacial colonization of brown trout Salmo trutta L. Ldh-5 as a phylogeographic marker. J Fish Biol, 35: 651–664.
Hansen, M. M., Loeschcke, V., Rasmussen, G. and Simonsen, V. (1993). Genetic differentiation among Danish brown trout (Salmo trutta) populations. Hereditas, 118: 177–185.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc, 58: 247–276.
Hindar, K., Jonsson, B., Ryman, N. and Ståhl, G. (1991). Genetic relationships among landlocked, resident, and anadromous brown trout Salmo trutta L. Heredity, 66: 83–91.
Hynes, R. A., Ferguson, A. and McCann, M. A. (1996). Variation in mitochondrial DNA and Postglacial colonization of north western Europe by brown trout. J Fish Biol, 48: 54–64.
Ibrahim, K. M., Nichols, R. A. and Hewitt, G. M. (1996). Spatial patterns of genetic variation generated by different forms of dispersal during range expansions. Heredity, 77: 282–291.
Karakousis, Y. and Triantaphyllidis, C. D. (1990). Genetic structure and differentiation among Greek brown trout (Salmo trutta L.) populations. Heredity, 64: 297–304.
Krieg, F. and Guyomard, R. (1985). Population genetics of French brown trout (Salmo trutta L.): large geographical differentiation of wild populations and high similarity of domesticated stocks. Génét Sél Evol, 17: 225–242.
Largiadìr, C. R. and Scholl, A. (1995). Effects of stocking on the genetic diversity of brown trout populations of the Adriatic and Danubian drainages in Switzerland. J Fish Biol, 47 (suppl. A), 209–225.
Largiadèr, C. R. and Scholl, A. (1996). Genetic introgression between native and introduced brown trout Salmo trutta L. populations in the Rhône River Basin. Mol Ecol, 5: 417–426.
Lessa, E. P. (1990). Multidimensional analysis of geographic structure. Syst Zool, 39: 242–252.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nei, M., Maruyama, T. and Chakraborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution, 29: 1–10.
Osinov, A. G. (1984). Zoogeographical origins of brown trout Salmo trutta (Salmonidae): data from biochemical genetic markers. J Ichthyol, 24: 10–21.
Osinov, A. G. and Bernatchez, L. (1996). ‘Atlantic’ and ‘Danubian’ phylogenetic groupings of brown trout Salmo trutta complex: Genetic divergence, evolution, and conservation. J Ichthyol, 36: 723–746.
Riffel, M., Storch, V. and Schreiber, A. (1995). Allozyme variability of brown trout (Salmo trutta L.) populations across the Rhenanian–Danubian watershed in southwest Germany. Heredity, 74: 241–249.
Ryman, N. (1983). Patterns of distribution of biochemical genetic variation in salmonids: differences between species. Aquaculture, 33: 1–21.
Skaala, Ø (1992). Genetic population structure of Norwegian brown trout. J Fish Biol, 41: 631–646.
Skaala, Ø and Jørdstad, K. E. (1987). Fine-spotted brown trout, its phenotypic description and biochemical genetic variation. Can J Fish Aquat Sci, 44: 1775–1779.
Skaala, Ø and Naevdal, G. (1989). Genetic differentiation between freshwater, resident and anadromous brown trout Salmo trutta within watercourses. J Fish Biol, 34: 597–605.
Sneath, P. H. A. and Sokal, R. R. (1973). Numerical Taxonomy. Freeman, San Francisco.
Sokal, R. R. (1991). The continental population structure of Europe. Ann Rev Anthropol, 20: 119–140.
Wright, S. (1978). Evolution and the Genetics of Populations, vol. 4, Variability Within and Among Natural Populations. University of Chicago Press, Chicago.
Acknowledgements
The authors are grateful to A. Osinov, A. Ferguson and an anonymous reviewer for useful discussion and criticism that enabled a clearer formulation of some of the concepts in this report. Stew Grant revised and improved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
García-Marín, JL., Utter, F. & Pla, C. Postglacial colonization of brown trout in Europe based on distribution of allozyme variants. Heredity 82, 46–56 (1999). https://doi.org/10.1038/sj.hdy.6884470
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884470
Keywords
This article is cited by
-
Large effective size as determinant of population persistence in Anostraca (Crustacea: Branchiopoda)
Conservation Genetics (2023)
-
Will the genomics revolution finally solve the Salmo systematics?
Hydrobiologia (2022)
-
Molecular characterization of spiny hedgehogs of the Iberian Peninsula: the missing link in the postglacial colonization of the western European hedgehog
Mammal Research (2021)
-
Brown trout (Salmo trutta L.) high genetic diversity around the Tyrrhenian Sea as revealed by nuclear and mitochondrial markers
Hydrobiologia (2019)
-
Molecular characterization and hybridization in Salmo (trutta) macrostigma morphotypes from Central Italy
Hydrobiologia (2013)