Abstract
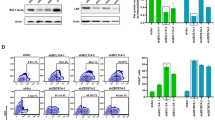
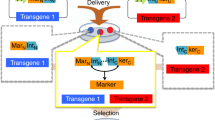
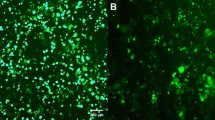
Vector-encoded fluorescent proteins (FPs) facilitate unambiguous identification or sorting of gene-modified cells by fluorescence-activated cell sorting (FACS). Exploiting this feature, we have recently developed lentiviral gene ontology (LeGO) vectors (www.LentiGO-Vectors.de) for multi-gene analysis in different target cells. In this study, we extend the LeGO principle by introducing 10 different drug-selectable FPs created by fusing one of the five selection marker (protecting against blasticidin, hygromycin, neomycin, puromycin and zeocin) and one of the five FP genes (Cerulean, eGFP, Venus, dTomato and mCherry). All tested fusion proteins allowed both fluorescence-mediated detection and drug-mediated selection of LeGO-transduced cells. Newly generated codon-optimized hygromycin- and neomycin-resistance genes showed improved expression as compared with their ancestors. New LeGO constructs were produced at titers >106 per ml (for non-concentrated supernatants). We show efficient combinatorial marking and selection of various cells, including mesenchymal stem cells, simultaneously transduced with different LeGO constructs. Inclusion of the cytomegalovirus early enhancer/chicken beta-actin promoter into LeGO vectors facilitated robust transgene expression in and selection of neural stem cells and their differentiated progeny. We suppose that the new drug-selectable markers combining advantages of FACS and drug selection are well suited for numerous applications and vector systems. Their inclusion into LeGO vectors opens new possibilities for (stem) cell tracking and functional multi-gene analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996; 272: 263–267.
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998; 72: 8463–8471.
Weber K, Bartsch U, Stocking C, Fehse B . A multi-color panel of novel lentiviral ‘gene ontology’ (LeGO) vectors for functional gene analysis. Mol Ther 2008; 16: 698–706.
Schüler A, Schwieger M, Engelmann A, Weber K, Horn S, Müller U et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood 2008; 111: 4532–4541.
Michaelis M, Rothweiler F, Klassert D, von Deimling A, Weber K, Fehse B et al. Reversal of P-glycoprotein-mediated multi-drug resistance by the MDM2 antagonist nutlin-3. Cancer Res 2009; 69: 416–421.
Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR . A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res 2009; 37: 1740–1754.
Kustikova OS, Wahlers A, Kühlcke K, Stähle B, Zander AR, Baum C, et al. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood 2003; 102: 3934–3937.
Fehse B, Kustikova OS, Bubenheim M, Baum C . Pois(s)on—it's a question of dose…. Gene Therapy 2004; 11: 879–881.
Rowitch DH . Glial specification in the vertebrate neural tube. Nat Rev Neurosci 2004; 5: 409–419.
Preuss E, Treschow A, Otte J, Brücher D, Felldin U, Alici E, et al. A novel, codon-optimised HSVtk(A168H) mutant for suicide gene therapy. Mol Ther 2009; 17: S223.
Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H . The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene 1991; 99: 249–254.
Lange C, Cakiroglu F, Spiess AN, Cappallo-Obermann H, Dierlamm J, Zander AR . Accelerated and safe expansion of human mesenchymal stromal cells in animal serum-free medium for transplantation and regenerative medicine. J Cell Physiol 2007; 213: 18–26.
Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 2005; 3: e283.
Niwa H, Yamamura K, Miyazaki J . Efficient selection for high expression transfectants with a novel eukaryotic vector. Gene 1991; 108: 193–199.
Blank U, Karlsson G, Karlsson S . Signaling pathways governing stem-cell fate. Blood 2008; 111: 492–503.
Mutschler M, Magin AS, Buerge M, Roelz R, Schanne DH, Will B et al. NF-E2 overexpression delays erythroid maturation and increases erythrocyte production. Br J Haematol 2009; 146: 203–217.
Uccelli A, Moretta L, Pistoia V . Mesenchymal stem cells in health and disease. Nat Rev Immunol 2008; 8: 726–736.
Ahn SM, Byun K, Kim D, Lee K, Yoo JS, Kim SU et al. Olig2-induced neural stem cell differentiation involves downregulation of Wnt signaling and induction of Dickkopf-1 expression. PLoS ONE 2008; 3: e3917.
Tey SK, Brenner MK . The continuing contribution of gene marking to cell and gene therapy. Mol Ther 2007; 15: 666–676.
Eilken HM, Nishikawa S, Schroeder T . Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 2009; 457: 896–900.
Taoudi S, Gonneau C, Moore K, Sheridan JM, Blackburn CC, Taylor E, et al., Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 2008; 3: 99–108.
Laker C, Meyer J, Schopen A, Friel J, Heberlein C, Ostertag W, et al. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol 1998; 72: 339–348.
Chambers I, Colby D, Robertson M, Nichols J, Lee S, Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003; 113: 643–655.
Ader M, Schachner M, Bartsch U . Transplantation of neural precursor cells into the dysmyelinated CNS of mutant mice deficient in the myelin-associated glycoprotein and Fyn tyrosine kinase. Eur J Neurosci 2001; 14: 561–566.
Ying QL, Smith AG . Defined conditions for neural commitment and differentiation. Methods Enzymol 2003; 365: 327–341.
Weber K, Fehse B . Diva-Fit: a step-by-step manual for generating high-resolution graphs and histogram overlays of flow cytometry data obtained with FACSDiva software. Cell Ther Transplant 2009; 1: doi 10.3205/ctt-2009-en-000045.01 (online published) .
Acknowledgements
We are grateful to Sabine Helbing for excellent technical assistance, Claudia Lange for providing MSC and Carol Stocking for critical reading of the paper. We thank many colleagues for their kind support with various cells and constructs: Luk van Parijs (pLentiLox3.7), Roger Y Tsien (tdTomato and mCherry cDNA), Atsushi Miyawaki/ Timm Schroeder (Venus cDNA), David W Piston (Cerulean cDNA), and David H Rowitch (murine Olig2 cDNA). This work was supported by the Deutsche Forschungsgemeinschaft (FE568/5-2; FE568/11–1 to BF), INTAS (06–1000013–9117 to BF), the Bundesministerium für Bildung und Forschung (BMBF 01GN0501 to UB), and the Frankfurter Stiftung für krebskranke Kinder. This report is part of the doctoral thesis of Kristoffer Weber.
Author contributions: KW conception and design, collection and/or assembly of data, data analysis and interpretation, and final approval of paper; UM collection and/or assembly of data and final approval of paper; BP collection and/or assembly of data and final approval of paper; UB collection and/or assembly of data and final approval of paper; and BF conception and design, data analysis and interpretation and paper writing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Weber, K., Mock, U., Petrowitz, B. et al. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther 17, 511–520 (2010). https://doi.org/10.1038/gt.2009.149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2009.149
Keywords
This article is cited by
-
Regulation of bone homeostasis by MERTK and TYRO3
Nature Communications (2022)
-
Mastering the use of cellular barcoding to explore cancer heterogeneity
Nature Reviews Cancer (2022)
-
Multicolor strategies for investigating clonal expansion and tissue plasticity
Cellular and Molecular Life Sciences (2022)
-
Integrin alpha-V is an important driver in pancreatic adenocarcinoma progression
Journal of Experimental & Clinical Cancer Research (2021)
-
Hematopoietic stem and progenitor cell proliferation and differentiation requires the trithorax protein Ash2l
Scientific Reports (2019)