Abstract
The activation of nuclear factor of activated T cells 5 (NFAT5), a well-known osmoprotective factor, can be induced by isotonic stimuli, such as activated Toll-like receptors (TLRs). It is unclear, however, how NFAT5 discriminates between isotonic and hypertonic stimuli. In this study we identified a novel context-dependent suppression of NFAT5 target gene expression in RAW 264.7 macrophages stimulated with lipopolysaccharide (LPS) or a high salt (NaCl) concentration. Although LPS and NaCl both used NFAT5 as a core transcription factor, these stimuli mutually inhibited distinct sets of NFAT5 targets within the cells. Although reactive oxygen species (ROS) are essential for this inhibition, the source of ROS differed depending on the context: mitochondria for high salt and xanthine oxidase for TLRs. Specifically, the high salt-induced suppression of interleukin-6 (IL-6) production was mediated through the ROS-induced inhibition of NFAT5 binding to the IL-6 promoter. The context-dependent inhibition of NFAT5 target gene expression was also confirmed in mouse spleen and kidney tissues that were cotreated with LPS and high salt. Taken together, our data suggest that ROS function as molecular sensors to discriminate between TLR ligation and osmotic stimuli in RAW 264.7 macrophages, directing NFAT5 activity toward proinflammatory or hypertonic responses in a context-dependent manner.
Similar content being viewed by others
Introduction
The nuclear factor of activated T cells (NFAT) family is a group of five transcription factors (NFAT1–5).1 NFAT5 was originally identified as a tonicity-regulated transcription factor involved in cellular protection from hypertonic stress.1 Accordingly, NFAT5 is highly expressed in tissues that are constantly exposed to osmotic stress, such as the kidney, intestinal epithelium and epidermis.2, 3 NFAT5 can also be activated under isotonic conditions, mediating physiologic or pathologic responses. For example, NFAT5 has a role in lymphocyte proliferation and survival,4 and mediates myoblast migration during skeletal muscle myogenesis.5 NFAT5 expression in bone marrow-derived cells also enhances atherosclerosis and drives macrophage migration.6 In addition, NFAT5-deficient mice show a marked reduction in antibody-induced arthritis, suggesting that NFAT5 has a key role in regulating rheumatoid arthritis.7
To date, it remains unclear how the transcription factor NFAT5 discriminates between isotonic and hypertonic stimuli, thereby producing context-dependent functional and molecular responses. This uncertainty may primarily be due to controversy surrounding the association between inflammation and osmotic stress. Several researchers have proposed that osmotic stress induced by a panoply of secreted proteins and proteolytic enzymes is the major cause of NFAT5 activation and subsequent inflammation.8 In contrast, we previously demonstrated that NFAT5 activity is increased by tumor necrosis factor-α and IL-1β stimulation under isotonic conditions.7 In addition, NFAT5 has been shown to contribute to innate immunity by activating gene expression in macrophages upon Toll-like receptor (TLR) ligation, independent of hypertonicity.9 Thus, NFAT5 may be part of a regulatory pathway that is distinct from the pathway induced by hypertonicity. Little is known, however, about the nature of this upstream signaling pathway under isotonic conditions or the pathway’s association with inter- or extracellular pathways under hypertonic conditions.
Despite the ubiquitous expression of NFAT5 in various tissues, this factor’s mechanism of action as a transcription factor is largely unknown. It has been proposed that most transcription factors perform specific functions, depending on the conditions under which the signaling pathway is activated.10 This context dependency is one of the major tools used by eukaryotes to expand on the complexity of transcriptional regulation through evolution.11 Molecular components of eukaryotic cells have continuously evolved toward multifunctionality through mutations and adaptations, several of which associate the components with a distinct signaling pathway.12 In these cases, the molecule will acquire a new function according to the upstream and downstream context of the signaling axis. The identification of the context to which a specific factor belongs will allow for the better selection of therapeutic targets and a greater understanding of the pathophysiology of the disease.
In this study we investigated the functional interaction between hypertonicity and TLR-induced NFAT5 signaling in RAW 264.7 macrophages in which both hypertonicity and inflammation were present. We found that a distinct source of reactive oxygen species (ROS) is required for the expression of NFAT5 target genes under hypertonic conditions compared with the source required for innate immunity: mitochondria under high-salt (NaCl) conditions and xanthine oxidase for TLRs. We also demonstrated how genes that respond to hypertonicity are affected by non-hypertonic inflammatory stimuli (or vice versa) and gained insight into the process by which NFAT5 achieves context-dependent activity. Collectively, our data provide new information on the ways in which cells acquire context dependency and display functional diversity.
Materials and methods
Reagents
The following antibodies were obtained from commercial sources: anti-nuclear matrix protein p84 (NMP p84; Abcam, Cambridge, MA, USA) and anti-α-tubulin (Sigma, St Louis, MO, USA). Anti-NFAT5 antibody was generated as previously described.13 Matrigel basement membrane matrix and DCFH-DA (2',7'-dichlorofluorescein diacetate) were purchased from BD Biosciences (San Jose, CA, USA), and the nuclear factor-κB reporter gene was obtained from Promega (Madison, WI, USA). All other reagents were purchased from Sigma.
Cloning of the NFAT5 reporter and IL-6 promoter reporter
To analyze the transcriptional activity of NFAT5, an NFAT5 consensus sequence with tandem repeats was transferred into the pEGFP-N1 vector (Clontech, Palo Alto, CA, USA) and the pDsRed-Express-N1 vector (Clontech), from which the cytomegalovirus promoter was then removed by the AseI and BamHI restriction enzymes. To construct a reporter system for the interleukin (IL-6) promoter, mouse genomic DNA, encompassing base-pair positions −938 to −11 relative to the start codon of the IL-6 gene, was cloned by PCR using the primers 5′-attaatcttcaacaacatgaggactgc-3′ and 5′-ggatccgaattgactatcgttctt-3′. The PCR product was subsequently subcloned into a pGEM-T Easy Vector System (Promega). The AseI/BamHI fragment containing the IL-6 promoter was then transferred to the pEGFP-N1 vector. Finally, the DNA fragment was confirmed by sequencing (Cosmo Genetech, Seoul, Korea).
Generation of stable cell lines
RAW 264.7 macrophages were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum in a humidified incubator at 37 °C. To construct a stable cell line transfected with the NFAT5 reporter gene, the cells were seeded to 50% confluence in 12-well plates and then transfected with green fluorescent protein (GFP)- or red fluorescent protein-fused NFAT5 reporter genes (5 μg) using Lipofectamine. After 3 days, the cells were reseeded to less than 20–25% confluence and were then selected by treatment with 125 μg ml−1 geneticin (Invitrogen, Carlsbad, CA, USA) for 3 weeks. Transfection rates were determined by fluorescence microscopy.
To produce NFAT5-deficient macrophages, RAW 264.7 macrophages were seeded to 20% confluence in 12-well plates and were then transduced with NFAT5 short hairpin RNA-harboring lentiviral particles (Santa Cruz Biotechnology, Santa Cruz, CA, USA) in the presence of polybrene (5 μg ml−1). Scrambled short hairpin RNA was used as a control. After 3 days, the cells were seeded to less than 20–25% confluence and selected by treatment with 5 μg ml−1 puromycin (Invitrogen) for 3 weeks. The level of knockdown was determined by immunoblotting for NFAT5 or by inspecting for fluorescent cells cotransfected with the GFP reporter by microscopy.
Flow cytometry
Analyses of reporter gene expression and ROS content in the intracellular space were performed using a FACSCanto flow cytometer (BD Biosciences, Heidelberg, Germany). Dead cells and debris were eliminated before analysis by forward- and side-scatter parameters. The fluorescence of the reporter gene and the presence of ROS were detected using GFP and fluorescein isothiocyanate filters, respectively. Changes in mean fluorescence intensity were determined using FlowJo analysis software (Tree Star, Ashland, OR, USA).
Fractionation and immunoblotting
To detect changes in NFAT5, cells were fractionated into cytosol and nuclei using hypo- and hypertonic lysis buffers, as previously described.7 Each lysate was separated on an 8% polyacrylamide gel containing SDS and then transferred to a nitrocellulose membrane. The membrane was agitated in blocking buffer containing anti-NFAT5, anti-NMP p84 and anti-α-tubulin antibodies overnight in a cold room and visualized using an enhanced chemiluminescence system. NMP p84 and α-tubulin were used as nuclear and cytosolic markers, respectively.
RNA isolation and real-time PCR
Total RNA was extracted from cells using an RNeasy Kit (Qiagen, Valencia, CA, USA). RNA samples from kidney tissues were prepared by homogenization using TRIzol reagent (Invitrogen), phenol–chloroform extraction and isopropanol precipitation. The RNA (1 μg per reaction) was reverse-transcribed to yield first-strand cDNA using reverse transcriptase (Takara, Shiga, Japan). To determine mRNA expression levels, cDNA converted from 1 μg of total RNA was diluted to various concentrations. The diluted cDNA was mixed with pairs of primers, including those primers for mouse aldose reductase (AR; 5′-agtgcgcattgctgagaactt-3′ and 5′-gtagctgagtagagtggccatgtc-3′), the betaine-γ-amino-butyric acid transporter (BGT-1; 5′-ctgggagagacgggttttgggtattacatc-3′ and 5′-ggaccccaggtcgtggat-3′), the sodium-dependent myo-inositol cotransporter (SMIT; 5′-ccgggcgctctatgacctggg-3′ and 5′-caaacagagaggcaccaatcg-3′), IL-6 (5′-ttccatccagttgccttcttg-3′ and 5′-aggtctgttgggagtggtatc-3′), and glyceraldehyde 3-phosphate dehydrogenase (5′-tgatgacatcaagaaggtggtgaa-3′ and 5′-tccttggaggccatgtaggccat-3′), along with cDNA sequences and SYBR Green PCR Master Mix (BD Biosciences), in a 20-μl total volume. The PCR cycling conditions were as follows: 5 min at 95 °C for 1 cycle, 30 s at 95 °C, and 1 min at 60 °C for 45 cycles. Gene expression was calculated by the comparative 2−ΔΔCt method after normalization to glyceraldehyde 3-phosphate dehydrogenase levels. The specificity of the amplification reactions was confirmed by melting curve analysis.
Enzyme-linked immunosorbent assays
Levels of IL-6 and MCP-1 in the culture supernatant were determined using an enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitationexperiments were performed according to the manufacturer’s recommendations (Millipore, Billerica, MA, USA). Briefly, cultured macrophages were fixed with 1% formaldehyde. The nuclei were isolated using fractionation buffer and sonicated three times on ice for 10 s to shear the DNA into 200–1000 bp fragments. A small aliquot (50 μl) was stored as input DNA. Chromatin-containing lysates were incubated with anti-NFAT5 antibody. DNA–protein immunocomplexes were precipitated with protein A agarose coated with salmon sperm DNA and then treated with proteinase K. The DNA samples were extracted with phenol/chloroform and precipitated with an ethanol and glycogen solution. To analyze NFAT5 enrichment within the IL-6 promoter, immunoprecipitated DNA samples were amplified by PCR using a primer pair for the IL-6 promoter (5′-agctttacgttctctttctcctta-3′ and 5′-aagtgactcagcacttgagca-3′), as described previously.9 The exon region of IL-6 (5′-ttccatccagttgccttcttg-3′ and 5′-aggtctgttgggagtggtatc-3′) was used as a negative control. Immunoprecipitated DNA and input DNA enrichments were subjected to 35 cycles of PCR (94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s) in 25 μl reaction mixtures. The PCR products then underwent electrophoresis on a 2% agarose gel in Tris-acetate-EDTA buffer and were visualized using SafeView reagent (Applied Biological Materials, Vancouver, BC, Canada).
Matrigel plug reporter assay
In vivo transcriptional activity was measured using the Matrigel plug reporter assay.14 Briefly, 8-week-old male BALB/c mice (Orient Bio, Seongnam, Korea) were subcutaneously injected in the dorsal region with 0.6 ml of Matrigel (BD Biosciences) containing 1 × 107 RAW 264.7 cells stably transfected with an red fluorescent protein-NFAT5 reporter. After stimulation of the cells with lipopolysaccharide (LPS) in vivo, the fluorescence intensity of each Matrigel complex was measured using a Maestro 2 Imaging System (CRi, Woburn, MA, USA). Images were normalized and analyzed using Maestro software 2.8 (CRi). Red fluorescent protein activity was quantified as the total photons per second per centimeter squared per steradian ( × 106 p s−1 cm−2 sr−1).
Statistical analysis
The data are expressed as the mean±s.d. Comparisons between groups were made using paired or unpaired Mann–Whitney U-tests. P-values less than 0.05 were considered to be statistically significant.
Results
Induction of NFAT5 activity by TLR compared with high salt concentration in macrophages
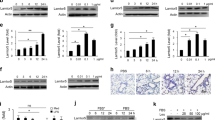
Using an NFAT5-specific GFP reporter construct, we confirmed that TLR ligation by LPS or heat-inactivated Escherichia coli was capable of inducing NFAT5-dependent reporter activity in RAW 264.7 macrophages (Figure 1a). These findings were consistent with the results of a previous report.9 As hypertonicity is a well-known stimulus for NFAT5 activation,1 we tested whether TLR ligation affects high salt-induced NFAT5 activation. When RAW 264.7 macrophages were cotreated with LPS (or E. coli) and NaCl, additive increases in NFAT5-dependent reporter activity were observed (Figure 1a). Furthermore, cotreatment with LPS and NaCl induced additive increases in the trafficking of NFAT5 from the cytoplasm to the nucleus in macrophages (Figure 1b). In accordance with the in vitro results, combined treatment resulted in a modest additive increase in NFAT5-dependent reporter activity in the in vivo Matrigel assay (Figure 1c). Taken together, these results suggest that TLR ligation does not suppress or sensitize high salt-induced NFAT5 expression or reporter activity in macrophages.
NFAT5 differentially affects nuclear factor-κB (NF-κB) activity in macrophages in a stimulus-dependent manner. (a and b) Additive effects of LPS and NaCl on NFAT5 activation. LPS (5 μg ml−1) or heat-inactivated E. coli (3 × 106 CFU ml−1) were added to RAW 264.7 cells for 24 h in the presence or absence of NaCl (90 mM). NFAT5-green fluorescent protein (GFP) activity and NFAT5 translocation were determined by flow cytometry and western blot analysis, respectively. The data on the right in a show the mean±s.d. of three independent experiments. *P<0.01 versus LPS or heat-inactivated E. coli alone. (c) LPS- and high salt-induced increases in NFAT5 reporter activity in vivo. Matrigel with RAW 264.7 macrophages stably transfected with NFAT5-red fluorescent protein (RFP) reporter were subcutaneously implanted into mice. On day 9, LPS (10 mg kg−1) and hypertonic saline (11.3% HS, 25 cc kg−1) were injected intraperitoneally. Normal saline (0.9% NS, 25 cc kg−1) was injected as a control. At 16 h post injection, NFAT5-RFP activity in the Matrigel was determined using the Maestro Imaging System. (d) Transcriptional activity of NF-κB in NFAT5-deficient macrophages treated with LPS (5 μg ml−1) or NaCl (90 mM). An NF-κB reporter containing GFP was transiently transfected into cells, and GFP activity representing NF-κB was determined by flow cytometry (left). The bar graph on the right represents the mean±s.d. of five independent experiments. *P<0.05 versus control short hairpin RNA (shRNA)-transfected cells in the presence or absence of NaCl. The reduction in NFAT5 expression was confirmed by western blot analysis (top panel).
It has been reported that under hyperosmolar conditions, NFAT5 modulates nuclear factor-κB activity in renal tubular epithelial cells.15 In our previous study, performed in RAW 264.7 cells, LPS did not increase tonicity-inducible genes, including AR, BGT-1 and SMIT, in contrast to high salt concentrations.16 Conversely, nitrite and IL-6 production were not altered by NaCl but were strongly increased by LPS.16 This finding suggests that the context-dependent activation of NFAT5 target genes is a function of the type of stimulus. In this study, NFAT5 short hairpin RNA did not affect nuclear factor-κB-dependent reporter activity in RAW 264.7 macrophages activated by LPS, which was mitigated by high-salt conditions (Figure 1d). These data, in conjunction with the findings of previous studies,15, 16 suggest that although both LPS and NaCl utilize NFAT5 as a key transcription factor for cell signaling, the downstream effects are propagated in distinct ways, depending on the nature of the stimulus.
Inhibition of NFAT5 target genes by LPS or high salt
A recent study demonstrated that NFAT5 regulates TLR-induced proinflammatory gene expression in macrophages, including the expression of IL-6, cyclooxygenase 2 and induced nitric oxide synthase, which is mediated by nuclear factor-κB activation.9 We also have shown that IL-6 production is nearly entirely dependent on NFAT5 activation following TLR ligation in RAW 264.7 macrophages.16 In this study we investigated the possibility of a functional interaction between LPS and hypertonicity-induced NFAT5 signaling by cotreating with LPS and NaCl. As observed in Figure 2a, we first confirmed the context-dependent activation of NFAT5 target genes in RAW 264.7 macrophages after individual treatment with NaCl or LPS. Interestingly, the expression of tonicity-inducible genes that are specifically controlled by NFAT5,17 including AR, BGT-1 and SMIT, was blocked by the pretreatment of macrophages with LPS (Figure 2a). Conversely, pretreatment with high salt inhibited the LPS-induced increase in IL-6 protein and mRNA expression (Figure 2b). This result was reproduced when the cells were simultaneously stimulated by LPS and NaCl (data not shown). Similarly, LPS-induced IL-6 promoter activity was also partially blocked by the cotreatment of cells with NaCl (Figure 2c). These results demonstrate that although LPS and hypertonicity share NFAT5 as a core transcription factor, these stimuli reciprocally inhibit the expression of downstream target genes.
Mutual suppression of NFAT5-governed gene expression by cotreatment with hypertonic stimuli and LPS. (a) Reduction in high salt-induced increases in AR, BGT1 and SMIT expression by pretreatment with LPS. RAW 264.7 macrophages were preconditioned with LPS (5 μg ml−1) for 4 h and then costimulated with NaCl (90 mM) for 4 h, or were stimulated first with NaCl for 4 h and then cotreated with LPS for an additional 4 h. The mRNA expression levels of AR, BGT1 and SMIT were assessed by real-time PCR. Fold inductions were calculated using the 2−ΔΔCt method. The data are expressed as the mean±s.d. of three independent experiments. *P<0.01. (b) Suppression of LPS-triggered IL-6 expression by cotreatment with NaCl. IL-6 expression levels were determined by enzyme-linked immunosorbent assay (left panel) and real-time PCR analysis (right panel). *P<0.01. (c) Hypertonic regulation of IL-6 promoter activity. RAW 264.7 cells were stimulated with LPS in the presence or absence of NaCl (90 mM) for 24 h and were subsequently subjected to flow cytometry analysis of IL-6 promoter activity. *P<0.01.
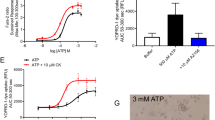
Mannitol is widely used in anesthesia, critical care medicine and in cases of sepsis.18 Although this drug’s clinical effects were originally attributed to the osmotic regulation of cells, other mechanisms, including anti-inflammatory action,19 have also been proposed. In this study we tested whether mannitol induces NFAT5 activity and alters TLR-triggered IL-6 expression in macrophages. Similar to LPS stimulation, mannitol treatment dose-dependently increased NFAT5-dependent reporter activity in RAW 264.7 macrophages (Figures 3a and b). Moreover, LPS-induced IL-6 production was partially inhibited by cotreatment with mannitol in a dose-dependent manner. Mannitol alone, however, did not affect IL-6 expression (Figure 3c). Thus, similar to high salt hyperosmolar mannitol increases NFAT5 activity and downregulates IL-6 production by macrophages costimulated with LPS.
Mannitol induces NFAT5 activity but reduces LPS-induced IL-6 production. (a and b) Dose-dependent increases in NFAT5 reporter activity after treatment with mannitol. Mannitol (100, 200 and 400 mM) was added to RAW 264.7 macrophages harboring the NFAT5 reporter gene for 24 h. The bar graph in b shows the mean±s.d. of three independent experiments. (c) Suppression of LPS-triggered IL-6 production by cotreatment with mannitol. RAW 264.7 macrophages were stimulated with LPS (5 μg ml−1) for 24 h in the presence of mannitol. IL-6 levels in the supernatant were determined by enzyme-linked immunosorbent assay. *P<0.01. The degree of cell viability was assessed by an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay and presented as the percentage of the cell viability of treated samples to the viability of control untreated cells.
ROS regulate context-dependent inhibition of NFAT5 target gene expression
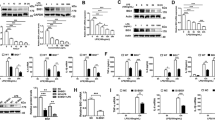
To date, the point at which LPS- and hypertonicity-mediated pathways diverge with respect to NFAT5 activation remains unknown. In addition, the mechanism by which LPS and NaCl reciprocally inhibit downstream targets is not well understood. We have shown that ROS regulate NFAT5 target gene expression in RAW 264.7 macrophages.16 Thus, we investigated whether ROS are involved in the context-dependent inhibition of NFAT5 target gene expression. Consistent with previous reports,20, 21 significant amounts of ROS were generated by RAW 264.7 macrophages that had been stimulated with either LPS or NaCl (Figure 4a). The time kinetics under the two conditions were similar, although LPS produced a greater quantity of ROS than did the high-salt condition (Figure 4a). When the cells were simultaneously treated with LPS and NaCl, ROS production was not suppressed (Figure 4b).
ROS regulate NFAT5 activity in a context-dependent manner. (a) Time kinetics of ROS production by RAW 264.7 macrophages stimulated with LPS or NaCl. ROS expression was determined by flow cytometry using an ROS probe (DCFH-DA (2',7'-dichlorofluorescein diacetate)). A representative histogram is shown on the left. The data on the right show the mean±s.d. of three independent experiments and are expressed as normalized mean fluorescence intensity (MFI) relative to the MFI determined 24 h after stimulation with 5 μg ml−1 LPS (set as 100%). *P<0.01 versus NaCl stimulation (90 mM). (b) Effects of LPS on high NaCl-induced ROS production by macrophages. ROS levels were determined 6 h after stimulation with 5 μg ml−1 LPS in the presence or absence of 90 mM NaCl. (c) Suppression of high salt-induced NFAT5 activity by myxothiazol (MTZ), a mitochondrial ROS inhibitor, but not by allopurinol (Allo). ROS scavengers, including Allo (1 mM) and MTZ (10 μM), were added to RAW 264.7 macrophages 1 h before stimulation with NaCl (90 mM) or LPS (5 μg ml−1). NFAT5-dependent reporter activity was then determined by flow cytometry. Representative histograms are shown on the left. The bar graph shows the mean±s.d. of three independent experiments.
Interestingly, in contrast to the hypertonic condition, allopurinol (a xanthine oxidase inhibitor) completely suppressed NFAT5 activation by LPS (Figure 4c), suggesting that xanthine oxidase is required for LPS-induced NFAT5 activation under nonhypertonic conditions. In the same cells, the induction of NFAT5 activity by high NaCl was not decreased by allopurinol (Figure 4c), indicating that xanthine oxidase regulation of NFAT5 activity is context dependent. Moreover, myxothiazol, which is an inhibitor of mitochondrial ROS,22 strongly inhibited high salt-induced SMIT mRNA expression in RAW 264.7 macrophages but did not downregulate LPS-induced IL-6 and nitric oxide synthase mRNA expression (Figures 5a and b). These data suggest that sources of ROS that mediate NFAT5-dependent increases in IL-6, AR and SMIT expression differ depending on context: mitochondria for high salt and xanthine oxidase for TLRs. In addition, we found that LPS-induced suppression of SMIT mRNA expression was completely restored by the addition of allopurinol (Figure 5c). In contrast, LPS-induced IL-6 production by macrophages was nearly completely recovered after the addition of rotenone and myxothiazol, which are inhibitors of mitochondrial complex-dependent ROS (Figure 5d). Collectively, these results suggest that the reciprocal inhibition of LPS and NaCl is achieved via xanthine oxidase-derived ROS and mitochondrial complex I-dependent ROS, respectively.
ROS control LPS- and high salt-induced suppression of NFAT5 target genes. (a and b) ROS dependency of NFAT5 target gene expression induced by LPS or high salt. The mRNA expression of AR, SMIT, IL-6 and NOS2 was determined by real-time PCR analysis 4 h after stimulation with NaCl or LPS. The data are the mean±s.d. of three independent experiments. *P<0.05 versus NaCl (A) or LPS alone (B). (c and d) ROS inhibitors restore SMIT, IL-6 and MCP-1 expression after combined treatment with high salt and LPS. RAW 264.7 macrophages were pretreated with various concentrations of allopurinol (Allo), rotenone (Ro), or myxothiazol (MTZ) for 1 h and then stimulated with LPS (5 μg ml−1) and NaCl (90 mM) for 4 h. IL-6 and MCP-1 concentrations in the culture supernatants were assessed by enzyme-linked immunosorbent assay. SMIT mRNA expression was determined by real-time PCR. *P<0.01. The degree of cell viability was assessed by an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay and presented as the percentage of the cell viability of treated samples to the viability of control untreated cells.
We and others have demonstrated that NFAT5 can directly bind to the IL-6 promoter and that this binding is enhanced by LPS stimulation.9, 16 In this study we used a chromatin immunoprecipitation assay with an anti-NFAT5 antibody to confirm the LPS-induced binding of NFAT5 to the IL-6 promoter (base pairs −938 to −11 relative to the start codon of the IL-6 gene) (Figure 6a). We then tested whether ROS directly modulated the interaction of NFAT5 with the IL-6 promoter. As observed in Figure 6b, allopurinol, but not rotenone, completely blocked NFAT5 binding to the IL-6 promoter. In addition, cotreatment with NaCl reduced NFAT5 binding to the IL-6 promoter in RAW 264.7 macrophages, a trend that was completely reversed by the addition of rotenone. These results suggest that ROS that are potentially mitochondrially derived inhibit NFAT5 binding to the IL-6 promoter, which mediates the high salt-induced suppression of IL-6 production.
Rotenone restores the high salt-induced suppression of NFAT5 binding to the IL-6 promoter. (a) Chromatin immunoprecipitation (ChIP) assay for the NFAT5 binding site within the IL-6 promoter. RAW 264.7 macrophages were stimulated with LPS (5 μg ml−1) for 10 h. DNA isolated from anti-NFAT5 precipitation complexes was amplified by PCR using specific primers for the promoter or exon regions of IL-6. The exon region of the IL-6 gene was used as a negative control. (b) RAW 264.7 macrophages were pretreated with allopurinol (Allo; 1 mM) or rotenone (Ro; 10 μM) for 1 h, stimulated with LPS (5 μg ml−1) and NaCl (90 mM) for 10 h, and then subjected to the ChIP assay. The PCR band densities were normalized relative to the input signals and quantified using ImageJ software (top panel). A representative of three independent experiments is shown. IP, immunoprecipitation.
We next tested whether the reciprocal inhibition of NFAT5 target genes could be reproduced using hyperosmolar stimuli and whether this inhibition could be accomplished in other cell types. As observed in Supplementary Figure 1a, hyperosmolar mannitol dose-dependently inhibited IL-6 production by RAW 264.7 macrophages, and this production was nearly completely restored by cotreatment with rotenone. Moreover, in renal tubular epithelial cells (TCMK-1 cells), LPS-induced IL-6 production was markedly reduced by the addition of LPS (Supplementary Figures 1b and c), whereas a high salt-induced increase in SMIT mRNA expression was only partially blocked by the addition of LPS (Supplementary Figure 1d). Combined treatment, with the addition of rotenone, dose-dependently recovered the high salt suppression of LPS-induced IL-6 production, although this effect was modest (Supplementary Figures 1b and c). Together, these results suggest that the context-dependent inhibition of NFAT5 activation and its dependency on ROS occur in multiple cell types, although to different extents, depending on the stimulus and cell type.
Context-dependent inhibition of NFAT5 target gene expression in vivo
We sought to confirm the in vitro findings of the context-dependent inhibition of NFAT5 target gene expression in the spleen and kidney of mice treated with LPS and high salt. As shown in Figure 7a, the administration of hypertonic saline, but not isotonic saline, suppressed LPS-induced IL-6 mRNA expression in the mouse spleen. In addition, the kidney cells of the mice cotreated with NaCl and LPS displayed a significant decrease in the expression of genes typically induced by high salt, such as SMIT and AR (Figures 7b and c). To summarize, we demonstrated that LPS and NaCl both mediated NFAT5 activation via ROS, but reciprocally suppressed the expression of NFAT5 downstream genes, including IL-6, AR and SMIT, both in vitro and in vivo (Figure 7d).
In vivo evidence for the suppression of NFAT5-governed genes by cotreatment with high salt and LPS. (a) High salt-induced suppression of IL-6 mRNA expression in the spleen. LPS (10 mg kg−1) was injected intraperitoneally into mice for 7 h after challenging the animals with hypertonic saline (HS; 11.3% HS, 25 cc kg−1) or normal saline (NS; 0.9% NS, 25 cc kg−1). mRNA expression of IL-6 in the spleen was analyzed by real-time PCR. *P<0.01. (b and c) LPS-mediated regulation of tonicity-responsive genes in the kidney. Under the same conditions as in a, the mRNA expression of IL-6, SMIT and AR in the kidney cells was analyzed by real-time PCR. *P<0.01. (d) Hypothetical model depicting the role of xanthine oxidase-derived ROS in the context-dependent activation of NFAT5, leading to IL-6 production upon TLR-4 ligation. ROS (XO), xanthine oxidase-derived ROS; ROS (M), mitochondrially generated ROS.
Discussion
The most widely observed method of expanding transcriptional regulation is by acquiring a new and distinct function that places the transcriptional machinery within a distinct signaling pathway.11 In fact, many transcription factors have been shown to perform a variety of functions, depending on the context of the cellular environment.23 For example, CREB and Tlx3 participate in cellular homeostasis and differentiation by executing context-dependent transcriptional regulation according to the nature of the stimulus.10, 12 In this study we found that high salt and TLR ligation activate distinct sets of downstream target genes in RAW 264.7 macrophages in an NFAT5-dependent manner. Although ROS are essential for this phenomenon, their sources differ depending on context: mitochondria for high salt and xanthine oxidase for TLRs. These two pathways are mutually suppressive both in vitro and in vivo. Moreover, ROS regulate the context-dependent inhibition of NFAT5 target gene expression by high salt or TLR activation. Specifically, high salt-induced suppression of IL-6 production is mediated through the ROS-induced inhibition of NFAT5 binding to the IL-6 promoter. These data provide intriguing evidence for the acquisition of context dependency.
ROS are generated by one of three mechanisms: (1) a xanthine oxidase-dependent, (2) a mitochondrial complex-dependent, or (3) an NADPH oxidase-dependent pathway.24 There are differences in the properties of each ROS type, with previous studies suggesting that whereas mitochondrially generated ROS are associated with the progression of heart failure, ROS derived from xanthine oxidase are involved in ischemic reperfusion and endothelial dysfunction.25, 26 We have also demonstrated that xanthine oxidase-induced ROS, but not mitochondria-derived ROS, have a key role in the progression of inflammatory arthritis by activating NFAT5.16
Demonstrating differences in the biochemical characteristics of mitochondria-derived and xanthine oxidase-induced ROS is a technically challenging process, mainly due to these species’ extremely short half-lives. Our results, however, may help to more easily distinguish between these properties. We presumed that the major source of ROS was likely different under hypertonic and nonhypertonic inflammatory conditions. We then observed that NFAT5-dependent IL-6 production was higher in RAW 264.7 macrophages treated with LPS alone than in cells cotreated with LPS and NaCl. In addition, decreased IL-6 promoter enrichment in cotreated cells was restored by pretreatment with the mitochondrial complex inhibitor rotenone. Conversely, LPS-induced suppression of SMIT, a tonicity-inducible gene, was completely reversed by xanthine oxidase inhibition. ROS from different sources may act directly on NFAT5 or act indirectly by initiating the differential activation of other signaling molecules within the cells. In fact, individual ROS with distinct chemical properties have been shown to be involved in specific signaling,27 displaying significant diversity in the types of affected transcription factors.28
Although it was previously established that hypertonic solution injections alleviate inflammatory responses and improve sepsis,29, 30 it was unclear how this effect was achieved. In the present study we demonstrated that pretreatment of RAW 264.7 macrophages with high salt or hyperosmolar mannitol blocked LPS-induced IL-6 production, which was recovered by cotreatment with rotenone. These results indicate that hypertonic regulation of the proinflammatory response is mediated by the context-dependent activation of NFAT5 target genes through xanthine oxidase- and/or mitochondrial complex I-derived ROS. Of note, when LPS was added to the macrophages in the presence of NaCl, NFAT5 and ROS production were not suppressed (Figures 1a and 4b). In addition, the time kinetics of the increases in NFAT5 and ROS did not differ between cells stimulated with LPS or NaCl (Figure 4a, data not shown). These results suggest that the quantitative changes in and time kinetics of NFAT5 and ROS are irrelevant to the context-dependent inhibition of IL-6 production and to the proinflammatory response induced by TLR ligation.
Although the context-dependent inhibition of NFAT5 activation and its dependency on ROS occur in multiple cell types, the extent seems to differ depending on the stimulus and cell type (Supplementary Figure 1). Moreover, several NFAT5 target genes were not completely dependent on ROS in RAW 264.7 macrophages (for example, AR expression in Figure 5a), indicating that in addition to ROS, other products, including proinflammatory cytokines and prostaglandins, may be directly involved in NFAT5 induction and the subsequent activation of NFAT5 target genes. In support of this notion, prostaglandin E2, which is known to be rapidly induced by LPS and high salt, stimulates the expression of osmoprotective genes, such as AR and SMIT, in MDCK cells and promotes survival under hypertonic conditions.31 Further study will be required to clarify this issue.
The context dependency of NFAT5 demonstrated in the present study has several potential clinical applications. It is well established that infections frequently occurring under hypertonic or hyperosmolar conditions, as observed in dehydration and diabetic hyperglycemia, tend to facilitate damage to the kidneys and other organs. The pathophysiology of this organ damage, however, remains unknown. We found that the induction of osmoprotective gene expression (for example, AR and SMIT) by high salt in the kidney was significantly hampered by TLR4 stimulation both in vitro and in vivo. This finding suggests that damage to the kidney tissues may be caused by the context-dependent inhibition of the osmoprotective action of NFAT5. This notion is supported by earlier reports showing that the inhibition of AR and SMIT expression causes renal injury in animal models.32, 33 Thus, ROS inhibitors may be potential therapeutic agents for preventing or treating renal injury under hypertonic or hyperosmolar conditions that are often observed in cases of bacterial infection.
In summary, we identified a novel context-dependent suppression of NFAT5 target gene expression in RAW 264.7 macrophages, which may facilitate NFAT5-induced activation of proinflammatory or hypertonic responses. Although LPS and NaCl both use NFAT5 as a core transcription factor, these stimuli mutually inhibit distinct sets of NFAT5 target genes via ROS derived from xanthine oxidase and the mitochondria, respectively. Our data provide intriguing evidence for cell-acquired context dependency and functional diversity via a single transcription factor. In addition, our findings present the possibility of developing ROS inhibitors as therapeutic agents for treating NFAT5-dependent renal injury and chronic inflammatory diseases, including diabetic nephropathy, atherosclerosis, hypertension and chronic arthritis.16
References
López-Rodríguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A . Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 2001; 15: 47–58.
López-Rodríguez C, Aramburu J, Rakeman AS, Rao A . NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA 1999; 96: 7214–7219.
Heo JI, Lee MS, Kim JH, Lee JS, Kim J, Park JB et al. The role of tonicity responsive enhancer sites in the transcriptional regulation of human hsp70-2 in response to hypertonic stress. Exp Mol Med 2006; 38: 295–301.
Trama J, Lu Q, Hawley RG, Ho SN . The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner. J Immunol 2000; 165: 4884–4894.
O’Connor RS, Mills ST, Jones KA, Ho SN, Pavlath GK . A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J Cell Sci 2007; 120: 149–159.
Halterman JA, Kwon HM, Leitinger N, Wamhoff BR . NFAT5 expression in bone marrow-derived cells enhances atherosclerosis and drives macrophage migration. Front Physiol 2012; 3: 313.
Yoon HJ, You S, Yoo SA, Kim NH, Kwon HM, Yoon CH et al. NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum 2011; 63: 1843–1852.
Schwartz L, Guais A, Pooya M, Abolhassani M . Is inflammation a consequence of extracellular hyperosmolarity? J Inflamm (Lond) 2009; 6: 21.
Buxadé M, Lunazzi G, Minguillon J, Iborra S, Berga-Bolanos R, Del Val M et al. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med 2012; 209: 379–393.
Lemberger T, Parkitna JR, Chai M, Schutz G, Engblom D . CREB has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. FASEB J 2008; 22: 2872–2879.
Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 2003; 20: 1377–1419.
Kondo T, Sheets PL, Zopf DA, Aloor HL, Cummins TR, Chan RJ et al. Tlx3 exerts context-dependent transcriptional regulation and promotes neuronal differentiation from embryonic stem cells. Proc Natl Acad Sci USA 2008; 105: 5780–5785.
Kim JA, Jeon US, Kwon MS, Lim SW, Kwon HM . Transcriptional activator TonE-binding protein in cellular protection and differentiation. Methods Enzymol 2007; 428: 253–267.
Yoo SA, Yoon HJ, Kim HS, Chae CB, De Falco S, Cho CS et al. Role of placenta growth factor and its receptor flt-1 in rheumatoid inflammation: a link between angiogenesis and inflammation. Arthritis Rheum 2009; 60: 345–354.
Roth I, Leroy V, Kwon HM, Martin PY, Feraille E, Hasler U . Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-kappaB activity. Mol Biol Cell 2010; 21: 3459–3474.
Kim NH, Hong BK, Kim WU . Xanthine oxidase-derived ROS direct context-dependent action of NFAT5 toward inflammatory response in macrophages. Arthritis Rheum 2012; 64 (supplement): 198.
Burg MB, Kwon ED, Kultz D . Regulation of gene expression by hypertonicity. Annu Rev Physiol 1997; 59: 437–455.
Huang GS, Shih CM, Wu CC, Hu MH, Tsai CS, Liaw WJ et al. Hypertonic saline, mannitol and hydroxyethyl starch preconditioning of platelets obtained from septic patients attenuates CD40 ligand expression in vitro. J Trauma 2010; 68: 331–336.
Cuschieri J, Gourlay D, Garcia I, Jelacic S, Maier RV . Hypertonic preconditioning inhibits macrophage responsiveness to endotoxin. J Immunol 2002; 168: 1389–1396.
Matsuzawa A, Saegusa K, Noguchi T, Sadamitsu C, Nishitoh H, Nagai S et al. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol 2005; 6: 587–592.
Yang T, Zhang A, Honeggar M, Kohan DE, Mizel D, Sanders K et al. Hypertonic induction of COX-2 in collecting duct cells by reactive oxygen species of mitochondrial origin. J Biol Chem 2005; 280: 34966–34973.
Kulisz A, Chen N, Chandel NS, Shao Z, Schumacker PT . Mitochondrial ROS initiate phosphorylation of p38 MAP kinase during hypoxia in cardiomyocytes. Am J Physiol Lung Cell Mol Physiol 2002; 282: L1324–L1329.
Fry CJ, Farnham PJ . Context-dependent transcriptional regulation. J Biol Chem 1999; 274: 29583–29586.
Shin MH, Moon YJ, Seo JE, Lee Y, Kim KH, Chung JH . Reactive oxygen species produced by NADPH oxidase, xanthine oxidase, and mitochondrial electron transport system mediate heat shock-induced MMP-1 and MMP-9 expression. Free Radic Biol Med 2008; 44: 635–645.
Tsutsui H . Oxidative stress in heart failure: the role of mitochondria. Intern Med 2001; 40: 1177–1182.
Spiekermann S, Landmesser U, Dikalov S, Bredt M, Gamez G, Tatge H et al. Electron spin resonance characterization of vascular xanthine and NAD(P)H oxidase activity in patients with coronary artery disease: relation to endothelium-dependent vasodilation. Circulation 2003; 107: 1383–1389.
von Harsdorf R, Li PF, Dietz R . Signaling pathways in reactive oxygen species-induced cardiomyocyte apoptosis. Circulation 1999; 99: 2934–2941.
Liu H, Colavitti R, Rovira II, Finkel T . Redox-dependent transcriptional regulation. Circ Res 2005; 97: 967–974.
Junger WG, Liu FC, Loomis WH, Hoyt DB . Hypertonic saline enhances cellular immune function. Circ Shock 1994; 42: 190–196.
Pascual JL, Khwaja KA, Ferri LE, Giannias B, Evans DC, Razek T et al. Hypertonic saline resuscitation attenuates neutrophil lung sequestration and transmigration by diminishing leukocyte-endothelial interactions in a two-hit model of hemorrhagic shock and infection. J Trauma 2003; 54: 121–132.
Neuhofer W, Steinert D, Fraek ML, Beck FX . Prostaglandin E2 stimulates expression of osmoprotective genes in MDCK cells and promotes survival under hypertonic conditions. J Physiol 2007; 583: 287–297.
Ho HT, Chung SK, Law JW, Ko BC, Tam SC, Brooks HL et al. Aldose reductase-deficient mice develop nephrogenic diabetes insipidus. Mol Cell Biol 2000; 20: 5840–5846.
Kitamura H, Yamauchi A, Sugiura T, Matsuoka Y, Horio M, Tohyama M et al. Inhibition of myo-inositol transport causes acute renal failure with selective medullary injury in the rat. Kidney Int 1998; 53: 146–153.
Acknowledgements
We thank all members of the Institute of Bone and Joint Diseases at the Catholic University of Korea. This work was supported by grants from the Korea Healthcare Technology R&D Project, the Ministry for Health, Welfare and Family Affairs (number A092258), and the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (R33-2008-000-10064-0 and 2009-0080087).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kim, NH., Hong, BK., Choi, S. et al. Reactive oxygen species regulate context-dependent inhibition of NFAT5 target genes. Exp Mol Med 45, e32 (2013). https://doi.org/10.1038/emm.2013.61
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2013.61
Keywords
This article is cited by
-
The TLR-2/TonEBP signaling pathway regulates 29-kDa fibronectin fragment-dependent expression of matrix metalloproteinases
Scientific Reports (2021)
-
Sargassum horneri methanol extract rescues C2C12 murine skeletal muscle cells from oxidative stress-induced cytotoxicity through Nrf2-mediated upregulation of heme oxygenase-1
BMC Complementary and Alternative Medicine (2015)