Abstract
The Alps are one of the most significant geographical barriers in Europe and several isolated Swiss and Italian valleys retain the distinctive Ladin and Romansch languages, alongside the modern majority of Italian and German languages. Linguistically, Ladin belongs to the Romance languages, but some studies on mitochondrial DNA (mtDNA) variation have suggested a major Middle Eastern component to their genealogical origin. Furthermore, an observed high degree of within-population diversity has been interpreted as reflecting long-standing differentiation from other European populations and the absence of a major bottleneck in Ladin population history. To explore these issues further, we examined Y chromosome and mtDNA variation in two samples of Ladin speakers, two samples of German speakers and one sample of metropolitan Italian speakers. Our results (1) indicate reduced diversity in the Ladin-speaking and isolated German-speaking populations when compared to a sample of metropolitan Italian speakers, (2) fail to identify haplotypes that are rare in other European populations that other researchers have identified, and (3) indicate different Middle Eastern components to Ladin ancestry in different localities. These new results, in combination with Bayesian estimation of demographic parameters of interest (population size, population growth rate, and Palaeolithic/Neolithic admixture proportions) and phylogeographic analysis, suggest that the Ladin groups under study are small genetically isolated populations (subject to strong genetic drift), having a predominantly European ancestry, and in one locality, may have a greater Palaeolithic component to that ancestry than their neighbours.
Similar content being viewed by others
Introduction
‘Ladins’ is the name given to groups totalling approximately 25 000 individuals that live in the Italian Alps which speak multiple variants of a shared Romance language (Ladin).1, 2 They are located in three valleys (Val Gardena, Val Badia, and Val Fassa) and a few other locations. The Ladins are of particular interest to molecular anthropologists because mitochondrial DNA (mtDNA) sequence data have been used to argue that they are genetically differentiated from both neighbouring and other European populations and are highly diverse in terms of within-population mtDNA sequence variation.3, 4, 5 Stenico et al3 and Stenico et al4 argued that the high internal diversity they identified is inconsistent with the view that isolation, low effective population size, and subsequent genetic drift had brought about Ladin genetic differentiation from neighbouring populations. Instead they postulated that the differences are the result of differing patterns of settlement in alpine versus non-alpine surrounding regions before the establishment of the Ladin language, followed by some degree of isolation but without any major population bottleneck events. Furthermore, based on comparisons of the distribution of mtDNA hyper variable segment 1 (HVS1) sequence haplotypes in Ladins with those in other populations, and specifically on the high incidence of mtDNA haplogroup T (previously known as haplogroup 2; 63, 25, and 36% in Val Gardena, Val Badia, and Val Fassa samples, respectively), which is relatively rare in Europe but common in the Middle Eastern, they proposed a large Middle Eastern component to Ladin origins. However, the mtDNA sequence data quality on which these conclusions were based has been questioned.6, 7, 8 Substitutions at key sites were found on multiple branches of the mtDNA HVS1 network, despite being rare in other European populations. These substitutions have been described as ‘phantom’ mutations6 and have been attributed to systematic sequencing errors.
In an attempt to address concerns about data quality, Vernesi et al5 collected and analysed a further sample (n=20) from Colle Santa Lucia (see Figure 1), one of the two locations from which the samples analysed by Stenico et al3 were collected. Vernesi et al5 did not report sequences from any re-sampling from Selva Val Gardena/Wolkenstein. Although Vernesi et al5 reported differences from the earlier Colle Santa Lucia set (Fisher's Exact Test for Population Differentiation:9 P=0.038, comparison made in this study) and accepted the possibility of sequencing errors, they nevertheless concluded that the principal conclusions reached by Stenico et al3 stood, that is ‘Ladin-speaking inhabitants of the Alps do display a high internal mitochondrial diversity, and that they are one of the main European mitochondrial outliers.’
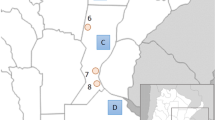
Map of the South Tyrol region in North Eastern Italy, showing the locations of the populations sampled for this study (LVG, LVB, GVU, GVL, ITA) as well as the location of Colle Santa Lucia (LSL) which was sampled by Stenico et al3 and by Vernesi et al5.GVL, German speakers from lower Vinschgau (Latsch) and surrounding area; GVU, German speakers from upper Vinschgau (Laas/Prad/Stilfs); ITA, Italian speakers from Bolzano and the surrounding area; LVB, Ladin speakers from Val Badia; LVG, Ladin speakers from Val Gardena.
Recently, Marroni et al10 investigated variation in Ladin and surrounding populations at a number of X-linked markers. They identified high linkage disequilibrium in Ladin samples and a high degree of differentiation both among different Ladin populations and between those populations and non-Ladin-speaking groups living in surrounding valleys. These results raise the possibility that Ladin differentiation is due to isolation and small population size, leading to high rates of genetic drift. Given the importance of the conclusions reached by Stenico et al3 and Stenico et al,4 involving prolonged isolation of a substantial population in south-central Europe displaying a Middle Eastern origin, we have investigated whether alternative explanations of Ladin demographic history are more plausible. In this study, we analysed mtDNA sequence data from the hypervariable region 1 (HVR1) of two further collections of Ladins and compared them with (a) neighbouring German (two sample sites) and Italian (one site) speakers to assess respective levels of genetic diversity and similarity and (b) multiple European and Middle Eastern populations (to assess the relative contributions of putative source populations to the five sample sets). We also undertake similar analyses of the paternally inherited non-recombining portion of the Y chromosome.
The South Tyrol: history, languages, and people
The South Tyrol region of the Alps contains a number of communities separated from one another by mountainous terrain. Three main languages; Italian, German, and Ladin, are spoken among them, together with a number of related dialects.2 Ladin, like Italian but unlike German, is a Romance language and, along with Romansch (spoken in eastern Switzerland) and Friulian (spoken in northeast Italy), belongs to the Rhaetian sub-group of the Italic branch of Indo-European languages. Although Ladin speakers alive today are mainly confined to three valleys, two of which are located in the South Tyrol region (Val Gardena and Val Badia), the distribution of toponyms (place names) suggests that Rhaetian languages were more widely spread during the early Middle Ages.11 The region in which the Ladin now live was incorporated into the Roman Empire only in around 15 or 16 BC (sometime after northern France (50 BC) but before Britain (43 AD)). German colonists are thought to have arrived following the fall of the Roman Empire in the 5th century AD.12 Today most non-Ladin-speaking groups in the South Tyrol, for which Italian is not their first language, speak German. The Italian speakers of the South Tyrol are found mainly in larger towns or cities, and their presence is largely the result of immigration encouraged by the Italian government after the detachment of South Tyrol from Austria in 1918, a practice that continued until the end of the Second World War.13
Materials and methods
Samples
Buccal swabs were collected from a total of 263 men from the South Tyrol region, comprising a sample of 59 Italian speakers from Bolzano and the surrounding area (ITA), 56 Ladin speakers from Val Badia (LVB), 46 Ladin speakers from Val Gardena (LVG), 52 German speakers from lower Vinschgau (Latsch) and surrounding area (GVL), and 50 German speakers from upper Vinschgau (Laas/Prad/Stilfs) (GVU) (see Figure 1). Donors were selected only if their grandfathers were from the same region and they were unrelated to other donors at the grandfather level. Swabs were stored in a DNA preservative solution containing 0.5% sodium dodecyl sulphate and 0.05 M ethylenediaminetetraacetic acid for transport purposes and DNA was purified by phenol–chloroform extraction/isopropanol precipitation. Samples were collected anonymously and informed consent was obtained from all individuals before samples were taken. To investigate Ladin variation in mtDNA, we also used the data published by Vernesi et al5 relating to a sample from the Ladin occupants of Colle Santa Lucia.
Molecular analysis
The mtDNA HVS-1 was sequenced as described previously.14 Sequences were obtained from all samples between positions 16008 and 16400 according to the numbering scheme of Anderson et al.15 mtDNA haplotypes were assigned to haplogroups (Mhg) for the purpose of admixture analysis only (see below) by identifying key combinations of HVS-1 mutations according to Macaulay et al,16 Richards et al,17 and Maca-Meyer et al18 as follows: 16069T, 16126C=Mhg-J, 16069T, 16126C, 16145A, 16261T=Mhg-J1, 16224C, 16311C=Mhg-K, 16126C, 16294T=Mhg-T, 16126C, 16163G, 16186T, 16189C, 16294T= Mhg-T1, 16126C, 16294T, 16304C=Mhg-T2, 16126C, 16256T, 16292T, 16294T=Mhg-T3, 16249C=Mhg-U1, 16051G=Mhg-U2, 16343G=Mhg-U3 16356C=Mhg-U4, 16270T=Mhg-U5, 16172C, 16219G=Mhg-U6, 16298C=Mhg-V, 16223T, 16292T=Mhg-W. For the remaining haplotypes, those with a T at position 16223 were assigned to Mhg-MNL and those with a C at position 16223 were assigned to Mhg-HVR.
Y chromosomes were typed in all samples for six microsatellites (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) and 11 Unique Event Polymorphism (UEP) markers (92R7, M9, M13, M17, M20, SRY+465, SRY4064, SRY10831, sY81, Tat, and YAP) as described previously.19 In addition, the UEP marker 12f2 was typed as described by Rosser et al.20 Microsatellite repeat numbers were assigned according to the nomenclature of Kayser et al.21 Y-chromosome haplogroups, defined by the 12 UEP markers, were classified according to the nomenclature proposed by the Y Chromosome Consortium.22 Their genealogical relationships are presented in Supplementary Figure S1.
Statistical and population genetic analysis
Unbiased genetic diversity (h) and its standard errors were calculated using the formulae given by Nei.23 Tests for the significance of differences in h-values were carried out using two methods. The first was a standard two-tailed z-test and the second was by bootstrap re-sampling. As a conservative measure, only the larger of the two P-values was used.14 To correct for multiple comparisons of h-values between different Ladin and non-Ladin population samples, we bootstrap re-sampled mtDNA and Y-chromosome haplotypes 10 000 times and calculated, in each case, the proportion of times that the average Ladin h-value was greater that the average non-Ladin h-value. As an conservative measure, we then multiplied the higher P-value by two as two tests were performed (one for mtDNA and one for Y chromosome). Tests for the significance of among-population differences in the within-population mean pairwise difference (MPD) for mtDNA sequences were carried out by bootstrap re-sampling 100 000 times. Populations were compared for differences in the distribution of haplotype and haplogroup frequencies using the genetic distance measure FST,24 and for differences in the distribution and molecular divergence of Y-chromosome microsatellite haplotypes using RST,25 estimated from Analysis of Molecular Variance ΦST values,26 and using the Fisher's Exact Test for Population Differentiation.9 The significance of pairwise FST and RST values was assessed by permuting haplotypes 10 000 times, and 10 000 Markov steps were used in the Fisher's Exact Test. All of the above analyses were performed using ARLEQUIN software.27 Patterns of genetic differentiation were visualized using classical multidimensional scaling, also known as Principal Coordinates Analysis,28 performed on a pairwise FST matrix, as implemented in the cmdscale command of the statistical package ‘R’ (URL: http://www.R-project.org/).
Admixture proportions were estimated using the likelihood-based method LEA29 [LEA can be obtained by contacting L Chikhi (e-mail: chikhi@cict.fr) or M Beaumont (e-mail: m.a.beaumont@reading.ac.uk)], based on Y-chromosome data alone, mtDNA data alone, and the Y-chromosome and mtDNA data sets combined. The combined analysis was performed under the assumptions of equal male/female demographic histories and equal effective population sizes. While these assumptions are unlikely to hold, the procedure provides gender-averaged estimates of demographic parameters that can be compared among populations. Since LEA does not model mutation and in our experience appears to have convergence problems when a large number (>30) of alleles are considered, only Y-chromosome haplogroup and inferred mtDNA haplogroup frequencies were used. Because no significant difference was observed between the two German-speaking populations (see Results and Table 1), we grouped these two samples together and treated them as a single population sample (GV). However, because the two Ladin samples were significantly different from one another in their distribution of mtDNA and Y-chromosome haplotypes using the Fisher's Exact Test of Sample Differentiation,9 we analysed them separately. We used unpublished Y-chromosome haplogroup frequency data for 179 Anatolian Turks (data available on request), as well as 57 Biscay Basques and 44 Guipuzcoa Basques.30 We combined this with published data from Rosser et al20 on 26 Basques and from Bosch et al31 and Perez-Lezaun et al32 on 50 Basques, giving comparable Y-chromosome haplogroup frequency data for 177 Basques. Because the same Y-chromosome UEP markers were not typed in all Basque samples, it was necessary to collapse haplogroups N3 and L (defined by the presence of derived states at loci Tat and M20 respectively) to their ancestral haplogroup, haplogroup K*(xL,N3,O2b,P) in both cases. Previously published mtDNA HVS-1 sequence data from Richards et al17 on 218 Anatolian Turks and 156 Basques was used. Most of the Anatolian Turk samples used by Richards et al17 are the same as those used here to generate Y-chromosome data. We ran 600 000 Monte Carlo iterations of the coalescent simulation and discarded the first 10 000 iterations as burn-in.
Demographic parameters of interest (population size and growth rate) were estimated from Y-chromosome data under a model of exponential growth (‘size model’ 1), using the Bayesian inference program BATWING (URL: http://www.maths.abdn.ac.uk/;ijw).33 We ran 102 000 Monte Carlo iterations of the coalescent simulation and discarded the first 2000 iterations as burn-in. Population demographic priors were: initial effective population size – gamma(1.1,0.0001), growth rate – gamma(1.01,1), time growth starts (in generations) – uniform(0, time to most recent common ancestor),34 final population size – uniform(0, 50 000). Locus-specific priors for the mutation rate per generation were based on observed mutations.34 These were: DYS19 – Gamma(3,1459), DYS390 – Gamma(5,929), DYS391 – Gamma(3,878), DYS392 – Gamma(2,878), and DYS393 – Gamma(1,878). As a precautionary measure, DYS388 was excluded from BATWING analysis because no published data on observed meioses are available for this locus. Population size and growth rate were estimated from mtDNA HVS1 sequence data analysed under a model of exponential growth using the Bayesian inference program BEAST v1.2, (URL: http://evolve.zoo.ox.ac.uk/beast/).35 Each population was analysed under a model of exponential growth, with an upper limit for population size of 50 000 and a mutation rate of 3.6 × 10−6 mutations/site/generation,17 under a HKY+G model of nucleotide substitution.36 We ran 200 000 000 Monte Carlo iterations to ensure that convergence had been achieved and that effective sample sizes were above a lower limit of 100 for all parameters. Post-processing of LEA, BATWING and BEAST outputs was carried out using the statistical package ‘R’ (URL: http://www.R-project.org/).
The geographic spread of South Tyrolean mtDNA haplotypes was quantified and visualized using the mtRadius database,8 currently containing data from over 24 000 individuals. A centre-of-gravity (COG) analysis37 was carried out using a minimal sequence range between positions 16093 and 16362,15 leaving a total of 19 493 individuals active in the database, of which approximately 12 000 were from Europe and surrounding areas. The method considers the worldwide geographic distribution of each sequence type individually, and optionally selects only those sequence types as reliable markers of origin which have a restricted geographic spread, measured as the mean distance of the haplotypes from their centre of gravity. Canary Islanders were excluded since they are mostly recent (post-1450s) immigrants from Spain, Italy, Britain, and elsewhere and would therefore obscure ancient geographic distributions rather than assist in describing them. This left a total of 17 917 individuals active in the database. COGs were calculated as described in Forster et al,37 and are based on at least two matches in the mtRadius database, with the frequency grid size set to 2 degrees of longitude by 2 degrees of latitude. As with the admixture analysis, because no differentiation was observed between the two German-speaking populations, we grouped the two samples (GV). However, because the two Ladin samples were significantly different from one another, we analysed them separately. For comparison, we also applied the COG analysis to 100 Anatolian mtDNA sequences, consisting of 50 randomly chosen Anatolian Turks and 50 randomly chosen Anatolian Kurds from mtRadius database, and removed those sequences from the active mtRadius database when that analysis was performed.
Results
mtDNA data quality
Because of previously highlighted concerns about data quality in published studies on Ladin mtDNA, we applied the methods suggested by Bandelt et al7 as a check on the quality of our mtDNA sequencing. We first restricted analysis of the data set of 263 individuals (all the South Tyrolean groups) to ‘weighty’ sites, as recommended by Bandelt et al7 (these are sites where the mutation rate appears relatively low). There were 40 such weighty sites. The ratio of weighty transition sites to transversion+indel sites (WTTI ratio) was 4.0 which is within the range reported for other European data sets considered by Bandelt et al7 to be reasonably problem-free. In addition, the reduced-median network drawn from the ‘weighty’ data contained five two-dimensional cycles and no higher-dimensional cycles, which is again within the range for reasonably problem-free data sets of this size.
mtDNA and Y-chromosome diversity
A total of 111 mtDNA HVS1 haplotypes were observed in the five population samples and these were clustered into 17 haplogroups based on the occurrence of key HVS-1 mutations (see Methods). The distribution of mtDNA haplotypes is given in Supplementary Table S1, together with the mtDNA haplogroup assignment. The Y-chromosome UEP markers defined seven observed haplogroups (HGs) (see Supplementary Figure S1 and Supplementary Table S2) and the UEP+microsatellite markers defined a total of 100 haplotypes (Supplementary Table S3). Two instances of homoplasy of microsatellite haplotypes across UEP haplogroups were observed (microsatellite haplotype 15 12 22 10 11 14 was found on two BR*(XDE, JR) and one E*(xE3a) chromosomes, and microsatellite haplotype 14 12 23 10 13 14 was found on one P*(xR1a) and one K*(xL,N3,O2b,P) chromosomes; all microsatellite haplotypes given in this paper are in the order DYS19, DYS388, DYS390, DYS391, DYS392, DYS393). This is consistent with other studies using similar numbers of microsatellite loci.38, 39, 40, 41, 42
To test for non-random association of mtDNA and Y-chromosome haplotypes among individuals within populations, which might indicate within group structuring or non-random sampling (such as sampling from multiple individuals within the same family), we carried out an extension of Fisher's Exact Tests to RxC tables.43 Because of the large number of mtDNA and Y-chromosome haplotypes present, all singleton mtDNA and Y-chromosome samples were respectively grouped into a single class. No significant association was found within any of the five populations.
Gene diversities were compared using P-values based on pairwise comparisons. mtDNA HVS1-haplotype frequency-based gene diversity values (see Supplementary Table S1) were significantly lower in the Ladins as a whole than in the Italian-speaking population (P<0.02), but not the German speakers when treated as a single population (P=0.16). Furthermore, in comparisons among the five populations sampled, the two Ladin groups had the lowest gene diversity with the Ladin Val Gardena group having a significantly lower h-value than the Italian speakers (P<0.005) and both German-speaking groups (P<0.04). All other pairwise population differences in h-values based on mtDNA HVS1-haplotype frequencies among the five groups sampled were greater than 0.05. By applying a correction for multiple tests to all five population samples, we found that the average Ladin gene diversity was lower than the average non-Ladin gene diversity in >99.99% of bootstrap re-sampled mtDNA haplotype data sets. Similarly, in both language and individual pairwise population group comparisons, the two Ladin samples displayed the lowest MPD for mtDNA HVS1 (Supplementary Table S4) although these differences were not significant when tested by bootstrap re-sampling.
Y-chromosome h-values (Supplementary Tables S2 and S3) were significantly lower in the Ladins than in the German or Italian-speaking populations at the levels of haplogroup frequencies (P<0.003) and UEP+microsatellite haplotype (‘full’ haplotype) frequencies (P<0.0003). A similar pattern was observed in pairwise comparisons of the five groups (P<0.05), except that the Val Gardena group was not significantly less diverse than the Lower Vinschgau group at the haplogroup (P=0.13). All other Y-chromosome pairwise population differences in h-values were greater than 0.05. Correcting for multiple tests among all five population samples (see above), we found that the average Ladin gene diversity was lower than the average non-Ladin gene diversity in 99.75% of bootstrap re-sampled Y-chromosome haplotype data sets.
Comparisons using various genetic distance measures (FST, RST, corrected mean pairwise nucleotide difference) and the Fisher's Exact Test of Sample Differentiation,9 at various levels of data (mtDNA sequence haplotypes, assigned mtDNA haplogroups, Y-chromosome full haplotypes, Y-chromosome microsatellite haplotypes and Y-chromosome UEP-defined haplogroups) indicated Ladin isolation. The ITA, GVU, and GVL groups are closely related to one another with the LVG and LVB groups more distantly related, both from one another and from the other three populations. While in pairwise comparisons among the ITA, GVU, and GVL samples, none of the genetic distance measures were statistically significant (P>0.05, unadjusted for multiple comparisons), genetic distances between the two Ladin populations and the ITA, GVU, and GVL populations were mostly significant and large. Table 1 shows FST values for population comparisons based on mtDNA HVS1-haplotype frequencies and Y-chromosome full haplotype frequencies. These distances are summarized in principal coordinate plots (Figures 2a and b). The Fisher's Exact Test of Population Differentiation, based on the same data, also indicates little differentiation among the ITA, GVU, and GVL samples (only the GVL/ITA groups mtDNA haplotype distribution was significantly different), while the LVG and LVB groups were significantly different from one another and the ITA, GVU, and GVL samples.
Estimation of current effective population size and growth rate
We estimated population demographic parameters of interest (current effective population size and population growth rate), based on mtDNA HVS1 sequence data using BEAST,35 and based on Y-chromosome data using BATWING.33 We analysed the two Ladin-speaking samples and the Italian-speaking sample individually but, because no differentiation was observed between the two German-speaking samples (see above and Table 1), we grouped them together and treated them as a single population (GV). Posterior estimates of current effective population size and population growth rate based on mtDNA and Y-chromosome data are given in Tables 2 and 3 respectively. To examine the difference in posterior estimates of these parameters in different populations, we took 200 000 two-point draws of the parameter value from its separate posterior distributions in populations #1 and #2, calculated the difference (value in population #2 − value in population #1) and calculated the proportion of instances that were greater than zero. For the analysis based on mtDNA data we found that 99.1% of GV, 99.0% of LVB, and 98.9% of LVG posterior estimates of current effective population size were smaller than those for ITA. We also found that 3.8% of GV, 8.6% of LVB, and 15.7% of LVG posterior estimates of population growth rate were smaller than those for ITA. For the analysis based on Y-chromosome data, we found that 99.3% of GV, 99.99% of LVB, and 99.9% of LVG posterior estimates of current effective population size were smaller than those for ITA, and 88.9% of GV, 90.2% of LVB, and 96.8% of LVG posterior estimates of population growth rate were smaller than those for ITA.
The origins of the Ladins: admixture analysis
Given the claim of a Middle Eastern origin for the Ladin speakers,4 we investigated whether there are differences among the five sample sets in the relative contributions from (a) to the east, Anatolian Turks, who have been used to represent Neolithic farmers, and (b) to the west, Basques, who have been used to represent Palaeolithic hunter-gatherers.44 To do so, we applied the admixture-based approach of Chikhi et al.29, 44, 45 While it is unlikely that these are the only possible source populations,46 they do represent convenient alternative options on which to assess comparative contributions to present day European populations.4 Posterior estimate modes of the Eastern/Neolithic contribution were similar for the German and Italian speakers when Y chromosome and mtDNA were considered together. However, while the LVG sample (mode=51%) was similar to the German and Italian speakers (mode=56%), the LVB sample (mode=13%) was very different. In comparisons with posterior estimates of Eastern/Neolithic contribution to the LVB sample 88.28, 92.56 and 92.15% of the estimates for the LVG, German speakers and the Italian speakers respectively were higher. Analysis based just on the Y chromosome produced a similar result. However, analysis of mtDNA alone revealed extreme estimates for the two Ladin-speaking populations with the LVB sample indicating the lowest contribution of all the groups (mode=18%) and the LVG sample the highest (mode=95%) (see Table 4 and Figure 3).
Estimated Middle Eastern admixture proportions (P1) for the LVB (solid lines), LVG (dashed lines), GV (alternately dotted and dashed lines), and Italian-speaking (dotted lines) populations based on inferred mtDNA haplogroup frequencies only (a) Y-chromosome haplogroup frequencies only (b) and both inferred mtDNA and observed Y-chromosome haplogroup frequencies combined (c). LVB, Ladin speakers from Val Badia; LVG, Ladin speakers from Val Gardena; mtDNA, mitochondrial DNA.
We also compared posterior estimates of the drift parameters T/N1, T/N2, and T/Nh indicating the amount of drift that has occurred in the Anatolian and Basque putative source populations and in the admixed populations, respectively, since admixture. Consistent with expectation based upon the timing of the acquisition of agriculture, all modal estimates of drift were higher for the Basques than for the Anatolian Turks. Consistent with isolation in both Ladin-speaking populations and the combined German-speaking population, all modal estimates of drift (T/Nh) were higher than those for the Italian-speaking population; for the combined Y-chromosome and mtDNA data sets, 88% of the Val Gardena, 82% of the Val Badia, and 94% for the German speaker posterior estimates of T/Nh were higher than for those for the Italian speakers. When mtDNA or Y-chromosome data were considered independently these values were 90, 91, 94, and 58%, 69, 59%, respectively.
Geographic centre-of-gravity analysis of mtDNA haplotypes
Applying the COG approach,37 the LVB, LVG, GV, and ITA haplotypes appear to be predominantly of recent European origin (see Supplementary Figure S2). Interestingly, a number of the Ladin speakers had their matching mtDNA haplotype COGs more clustered in the region of the Alps than did either the German or Italian speakers. Only six out of 102 Ladin speakers, three out of 102 German speakers, and one out of 59 Italian speakers have their COGs outside Europe. These outliers are likely to be due to parallel mutations in the short HVS1 sequences considered here and would be expected to disappear with longer sequences. For comparison, Supplementary Figure S2 shows a COG plot for 100 randomly selected Anatolian mtDNA sequences. It should be noted that the data for non-European populations are limited and, as such, these analyses will be biased to an unknown extent.
Discussion
Data analysed in this study support different conclusions from those reached by Stenico et al,3 Stenico et al,4 and Vernesi et al.5 There is little sharing of haplotypes between the Ladin samples included here and those presented previously (using only data for the region of sequence overlap – positions 16051–16362). Only two out of 18 haplotypes reported by Stenico et al3 were found among the 40 Ladin haplotypes reported here. In the case of the Vernesi et al5 study, 5 of the 10 haplotypes reported in their paper were found among the 40 Ladin haplotypes reported here (see Supplementary Table S1). In addition, the modal mtDNA HVS-1 sequence in our Ladin sample, the Cambridge Reference Sequence,15 was not found in any of the previously published Ladin data sets. Likewise, the modal mtDNA HVS1 sequence in the data set reported by Vernesi et al5 was found in only two of the 56 LVB individuals and none of the 46 LVG individuals. For comparison, 11 of the 40 Ladin haplotypes reported here were also found in our German- and Italian-speaking samples. Despite the small sample numbers reported in previously published studies, all possible Ladin comparisons, except that of Stenico et al's3 Santa Licia and Val Gardena samples, indicated significant sample differentiation by the Fisher's Exact Test for Population Differentiation.9 A second difference from the previous studies is that we find lower diversity in the Ladins than in their neighbouring populations, both in terms of gene diversity (h) and within-population MPD. When comparing all five populations investigated here, we found that the average Ladin gene diversity was lower than the average non-Ladin gene diversity in 99.75% bootstrap re-sampled data sets. In part, these differences may arise because only one location (Val Gardena) was sampled in both this and the previous three studies for which data are available. Despite the small sample size of the Stenico et al3 data (n=10), gene diversity for our Val Gardena sample is significantly lower (P=0.015) (using only data for the region of sequence overlap—positions 16051–16362). A third difference is that geographic analysis of the global distribution of mtDNA HVS1 haplotypes presented here using the COG approach and the mtRadius database37 does not indicate an unusually large Middle Eastern component to Ladin origins; nearly every Ladin mtDNA type has its closest genetic match in Europe rather than elsewhere. Of the few non-European outliers, both in the Ladin as well as in the Italian and German mtDNA samples, none cluster in the Near East and are best explained as chance matches caused by parallel mutations in the short sequence range considered. A potentially interesting feature distinguishing Ladin from German and Italian mtDNA is that three geographically specific Ladin types (found in 8 out of 102 Ladin individuals) are not common beyond the Alps. The German and Italian sequences do not demonstrate such clustering. In contrast, the plot generated for 100 randomly selected Anatolian mtDNA sequences shows that most haplotypes with a limited range (as indicated by black circles in Supplementary Figure S2) have their COGs in the Middle East. However, as discussed below, it should be noted that while admixture analysis does not indicate a high Middle Eastern component to Ladin ancestry overall, the result for the Val Gardena mtDNA data does suggest an unusually high contribution (mode=95%).
Significantly lower Y-chromosome diversity is also observed in the Ladins when compared to the neighbouring populations analysed here, and the average Ladin gene diversity was lower than the average non-Ladin gene diversity in >99.99% bootstrap re-sampled data sets. Furthermore, we find a relatively high degree of mtDNA and Y-chromosome differentiation, quantified using FST, both between the two different Ladin localities and also between the Ladin localities and the neighbouring Italian-speaking and two German-speaking populations. The most likely explanation for these observations is population isolation and a relatively small effective population size, leading to strong genetic drift in the Ladins. This is further supported by Bayesian inference of effective population size based on mtDNA HVS1 sequence data using BEAST, Y-chromosome data using BATWING, and by LEA analysis, which all show that posterior estimates of the effective population size are smaller (in the case of BEAST and BATWING analysis, by wide credible margins) for the Ladins than for the urban Italian speakers. The two German-speaking samples also have smaller posterior estimates of effective population size than the Italian speakers. As both the Ladin speakers and the German speakers were collected from isolated valleys, our study supports the contention that the Alps present geographic barriers that even today are reflected in the genetic make-up of the populations living there. This finding has also been noted in other mountainous regions (for example in mountainous parts of Armenia42).
The Ladin speakers sampled by Vernesi and colleagues5 were from a different location (Colle Santa Lucia) to those sampled here. Considering that those sequences were produced partly in response to a criticism of data quality,7 it seems unlikely that these data are also erroneous. While the high diversity observed in the Vernesi data set is not consistent with our general conclusion of high rates of genetic drift in Ladin groups, the difference in distribution of mtDNA haplotypes between their Ladin sample and those presented here is consistent with isolation among geographically separated populations. It is possible that the very different conclusions drawn by our analyses are due to genuine differences in the population history of Colle Santa Lucia. However, the fact that we find significant differences in the one Ladin group common to our study and that of Stenico et al3 (Val Gardena), plus the existence of other evidence for sequencing errors in those data (Bandelt et al7), strongly suggest that, with respect to the earlier Stenico et al3 paper, differences are due to laboratory-based artefacts.
Isolation and genetic drift will confound the task of identifying potential source populations, particularly when only a small number of loci are analysed. When two potential source populations are proposed, admixture analysis can be useful for estimating their relative contributions to the admixed population. Both Chikhi et al44 and Belle et al45 have used this approach to provide an estimate of the relative contributions of Neolithic farmers and Palaeolithic hunter-gatherers to various populations in Europe, using Near Easterners and Basques, respectively, as the descendents of their model source populations. Their chosen admixture estimation method29 has the advantage of allowing for, and providing an estimate of, the extent of drift in the admixed population and both source populations subsequent to the (single) admixture event. Our results are intriguing in that when mtDNA and Y-chromosome data are considered together, they suggest a larger Palaeolithic component to LVB ancestry than any of their neighbouring populations LVG, GV, and ITA. Examination of admixture proportions based on individual loci suggests that in this case the signal is mainly due to the Y-chromosome data. It is tempting to postulate that a low Neolithic component to LVB ancestry is the result of long-term isolation, with that population perhaps descending from an in situ Palaeolithic population. However, we are cautious of placing too much emphasis on the absolute estimates resulting from admixture analysis for the following reasons: (1) The method assumes the correct identification of source populations that have not been influenced by admixture themselves since the Neolithic, (2) different effective population size and demography for mtDNA and Y-chromosome data would violate the modelling assumptions of LEA when these loci are analysed together, (3) the admixture estimation method employed cannot account for mutation since the admixture event, (4) estimates of admixture are typically wide and overlapping for the four South Tyrolean populations, (5) it is highly unlikely that any European population formed following a single admixture event between incoming farmers and Palaeolithic hunter-gatherers. Of particular concern is the appropriateness of the source populations used, especially since Alonso et al46 have recently questioned whether Basques best represent Palaeolithic Europeans. Nonetheless, the admixture approach, using Near Easterners and Basques as source populations, has made an important contribution to the ongoing debate of the origins of Europeans44, 45 and, in the context of this study, serves to make useful comparisons between the various South Tyrolean groups. We note that for the LVG population, the mtDNA and Y-chromosome admixture estimates are rather discrepant; when mtDNA data are considered alone admixture estimates suggest a higher Neolithic/Middle Eastern component to LVG ancestry than any of their neighbouring populations. This is consistent with the findings of Stenico et al3, 4 but in contrast to the results of the COG approach using the mtRadius database37 and the admixture estimates based on Y-chromosome data alone. However, such a discrepancy is within the range of expectation in a small population experiencing high rates of genetic drift.
The high degree of differentiation between the two Ladin populations sampled here is intriguing in that it suggests a low level of gene flow between different localities, despite shared cultural and linguistic features. Genetic isolation among different Ladin populations is consistent with recent studies of X-chromosome variation10 as well as the highly tree-like phylogeny of regional Ladin languages, which indicate a language fissioning process with little subsequent vocabulary exchange.1 Perhaps, the best explanation for this is that geography is the most important isolating factor for the Ladins, and possibly the two German-speaking populations. However, we cannot rule out other processes such as local language replacement and admixture from other source populations. If the patterns of mtDNA and Y-chromosome variation observed in the South Tyrolean populations do reflect long-term isolation and a high rate of genetic drift, then this would explain the high levels of linkage disequilibrium recently observed.10 This could be potentially relevant to medical genetic research.
References
Forster P, Toth A, Bandelt H-J : Evolutionary network analysis of word lists: visualising the relationships between alpine romance languages. J Quant Ling 1998; 5: 174–187.
Ruhlen M : A Guide to The World's Languages. Stanford, CA: Stanford University Press, 1987.
Stenico M, Nigro L, Bertorelle G et al: High mitochondrial sequence diversity in linguistic isolates of the Alps. Am J Hum Genet 1996; 59: 1363–1375.
Stenico M, Nigro L, Barbujani G : Mitochondrial lineages in Ladin-speaking communities of the eastern Alps. Proc R Soc Lond B Biol Sci 1998; 265: 555–561.
Vernesi C, Fuselli S, Castri L, Bertorelle G, Barbujani G : Mitochondrial diversity in linguistic isolates of the Alps: a reappraisal. Hum Biol 2002; 74: 725–730.
Bandelt HJ, Lahermo P, Richards M, Macaulay V : Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 2001; 115: 64–69.
Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V : The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 2002; 71: 1150–1160.
Rohl A, Brinkmann B, Forster L, Forster P : An annotated mtDNA database. Int J Legal Med 2001; 115: 29–39.
Raymond M, Rousset F : An exact test for population differentiation. Evolution 1995; 49: 1280–1283.
Marroni F, Pichler I, De Grandi A et al: Population isolates in South tyrol and their value for genetic dissection of complex diseases. Ann Hum Genet 2006; 70: 812–821.
Pellegrini GB : La genesi del Retoromanzo (o Ladino). Tübingen: Niemeyer, 1991.
Leitner W : Die Urzeit; In: Fontana J, (ed).: Geschichte des Landes Tirol. Innsbruck-Wien-Bozen: Athesia-Tyrolia, 1985, vol 1.
Parteli O : 1918–1970; In: Fontana J, (ed).: Geschichte des Landes Tirol. Innsbruck-Wien-Bozen: Athesia-Tyrolia, 1985, vol 4.
Thomas MG, Weale ME, Jones AL et al: Founding mothers of Jewish communities: geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet 2002; 70: 1411–1420.
Anderson S, Bankier AT, Barrell BG et al: Sequence and organization of the human mitochondrial genome. Nature 1981; 290: 457–465.
Macaulay V, Richards M, Hickey E et al: The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 1999; 64: 232–249.
Richards M, Macaulay V, Hickey E et al: Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 2000; 67: 1251–1276.
Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM : Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2001; 2: 13.
Thomas MG, Bradman N, Flinn HM : High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 1999; 105: 577–581.
Rosser ZH, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Kayser M, Caglia A, Corach D et al: Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 1997; 110: 125–133, 141–129.
The Y Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Nei M : Molecular Evolutionary Genetics. New York: Columbia University Press, 1987.
Reynolds J, Weir BS, Cockerham CC : Estimation for the coancestry coefficient: basis for a short-term genetic distance. Genetics 1983; 105: 767–779.
Slatkin M : A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995; 139: 457–462.
Excoffier L, Smouse PE, Quattro JM : Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992; 131: 479–491.
Schneider S, Roessli D, Excoffier L : Arlequin ver 2.000: A Software for Population Genetics Data Analysis. Geneva, Genetics and Biometry Laboratory, University of Geneva: Switzerland, 2000.
Gower JC : Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1966; 53: 325–328.
Chikhi L, Bruford MW, Beaumont MA : Estimation of admixture proportions: a likelihood-based approach using Markov chain Monte Carlo. Genetics 2001; 158: 1347–1362.
Capelli C, Redhead N, Abernethy JK et al: A Y chromosome census of the British Isles. Curr Biol 2003; 13: 979–984.
Bosch E, Calafell F, Santos FR et al: Variation in short tandem repeats is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 1999; 65: 1623–1638.
Perez-Lezaun A, Calafell F, Seielstad M et al: Population genetics of Y-chromosome short tandem repeats in humans. J Mol Evol 1997; 45: 265–270.
Wilson IJ, Weale ME, Balding DJ : Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J R Statist Soc A 2003; 166: 155–201.
Weale ME, Weiss DA, Jager RF, Bradman N, Thomas MG : Y chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol 2002; 19: 1008–1021.
Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W : Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 2002; 161: 1307–1320.
Hasegawa M, Kishino H, Yano T : Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 1985; 22: 160–174.
Forster P, Calì F, Röhl A et al: Continental and subcontinental distributions of mtDNA control region types. Int J Legal Med 2002; 116: 99–108.
Bradman N, Thomas MG, Goldstein DB : The genetic origins of Old Testament priests; In: Renfrew C (ed).: America Past, America Present: Genes and Languages in the Americas and Beyond. Oxford, England: Oxbow, 2000, pp 31–44.
Malaspina P, Cruciani F, Ciminelli BM et al: Network analyses of Y-chromosomal types in Europe, northern Africa, and western Asia reveal specific patterns of geographic distribution. Am J Hum Genet 1998; 63: 847–860.
Scozzari R, Cruciani F, Santolamazza P et al: Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 1999; 65: 829–846.
Thomas MG, Parfitt T, Weiss DA et al: Y chromosomes traveling south: the cohen modal haplotype and the origins of the Lemba—the ‘Black Jews of Southern Africa’. Am J Hum Genet 2000; 66: 674–686.
Weale ME, Yepiskoposyan L, Jager RF et al: Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet 2001; 109: 659–674.
Mehta C, Patel N : Algorithm 643. FEXACT: A Fortran subroutine for Fisher's exact test on unordered r*c contingency tables. ACM Trans Math Software 1986; 12: 154–161.
Chikhi L, Nichols RA, Barbujani G, Beaumont MA : Y genetic data support the Neolithic demic diffusion model. Proc Natl Acad Sci USA 2002; 99: 11008–11013.
Belle EM, Landry PA, Barbujani G : Origins and evolution of the Europeans' genome: evidence from multiple microsatellite loci. Proc Biol Sci 2006; 273: 1595–1602.
Alonso S, Flores C, Cabrera V et al: The place of the Basques in the European Y-chromosome diversity landscape. Eur J Hum Genet 2005; 13: 1293–1302.
Acknowledgements
We acknowledge the generous help of Drs Pizzecco, Scherer, Skocir, Wallnöfer (Val Venosta), Capone, Corradini, Fontanella (Bolzano), Kostner, Troebinger (Val Gardena), and Mahlknecht (Val Badia) for collecting the buccal swabs. We thank HJ Bandelt for comments on early drafts of the manuscript and advice on assessing mtDNA sequence data quality. We also thank Drs A Drummond and A Rambaut for providing advice on the use of the BEAST software, and the two anonymous referees for their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Thomas, M., Barnes, I., Weale, M. et al. New genetic evidence supports isolation and drift in the Ladin communities of the South Tyrolean Alps but not an ancient origin in the Middle East. Eur J Hum Genet 16, 124–134 (2008). https://doi.org/10.1038/sj.ejhg.5201906
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201906
Keywords
This article is cited by
-
Mitogenomic diversity and stable isotopes provide insights into the maternal genetic history, mobility patterns, and diet of early medieval individuals from the Eastern Italian Alps
Archaeological and Anthropological Sciences (2022)
-
Evolution of morphological crypsis in the Tetramorium caespitum ant species complex (Hymenoptera: Formicidae)
Scientific Reports (2018)
-
Overcoming the dichotomy between open and isolated populations using genomic data from a large European dataset
Scientific Reports (2017)
-
Genetic diversity of Italian goat breeds assessed with a medium-density SNP chip
Genetics Selection Evolution (2015)
-
Interdisciplinary approach to the demography of Jamaica
BMC Evolutionary Biology (2012)