Abstract
The identification of subtelomeric rearrangements as a cause of mental retardation has made a considerable contribution to diagnosing patients with mental retardation. It is remarkable that for certain subtelomeric regions, deletions have hardly ever been reported so far. All the laboratories from the ‘Association des Cytogénéticiens de Langue Française’ were surveyed for cases where an abnormality of the subtelomere FISH analysis had been ascertained. Among 1511 cases referred owing to unexplained mental retardation, 115 (7.6%) patients showed a clinically significant subtelomeric abnormality. We report the clinical features and the molecular cytogenetic delineation of isolated de novo deletions on 20q13.33 in two cases. Detailed mapping was performed by micro-array CGH in one patient and confirmed by FISH in the two patients. We compare our data with the only three patients reported in the literature. Both patients shared a deleted region of approximately 1.33 Mb including 40 genes, with a 324 kb difference between the two patients. Haploinsufficiency for CHRNA4 and ARFGAP1 may have contributed towards a severe phenotype. In addition, the data in all patients suggest that haploinsufficiency for SOX18 may not cause the hypotrichosis–lymphedema–telangiectasia syndrome, or causes milder disease. Our study gives important information by defining the size of imbalance and better predicting the phenotype. Two clinically distinct phenotypes may be drawn, a mild mental retardation or a more complex and severe phenotype, according to the presence or absence of the CHRNA4 and ARFGAP1 genes respectively.
Similar content being viewed by others
Introduction
Cryptic subtelomeric rearrangements are estimated to account for approximately 5% of mental retardation (MR)/malformation syndromes, with a higher deletion rate in those with moderate to severe MR than in mild MR.1 Although in some subtelomeric rearrangements, the phenotype is well described, there are many cryptic rearrangements for which there is little or no phenotypic information complicating the diagnostic and prognostic implications. Therefore, further studies are needed to delineate genotype–phenotype correlations for each human subtelomeric region. All the laboratories from the ‘Association des Cytogénéticiens de Langue Française’ (http://www.eaclf.org) offering an appropriate cytogenetic service were surveyed over a 10-year period (1994–2004) for cases where a submicroscopic telomeric abnormality had been ascertained. Among 1511 cases referred owing to unexplained MR, 115 (7.6%) patients showed a clinically significant subtelomeric abnormality. We report the phenotypic and the molecular cytogenetic characterization of two patients with uncommon isolated de novo subtelomeric 20q deletions. Detailed mapping was performed by micro-array CGH in one patient and confirmed by FISH in the two patients. We compare our data with the only three previously reported cases in the literature. These findings are discussed with respect to the genes in the deleted region of chromosome 20q13.33.
Materials and methods
Patients
Patient 1 is the only child of unrelated parents, aged 35 (mother) and 34 (father). Family history was negative for congenital anomalies and/or MR. This girl was born at 38 weeks of gestation after an uneventful pregnancy and delivery. The birth weight, length and head circumference were normal. She sat at 13 months and walked at 19 months. At 7 years 10 months, she had mild psychomotor retardation associated with a nystagmus and her language abilities were mildly retarded with phonological abnormalities. A physical examination showed the length to be 124.5 cm (+0.3SD), weight 23 kg (–0.5SD) and head circumference 48.8 cm (–2.3SD). Craniofacial dysmorphism included microcephaly, brachycephaly, prominent metopic suture, temporal narrowing, mild hypertelorism, upslanting palpebral fissures, anteverted nares, down-turned corners of the mouth and a thin upper lip (Figure 1a). The neurological examination was normal. Audiogram, electroencephalogram (EEG), visual evoked potentials (VEP), electroretinogram (ERG), and a magnetic resonance imaging (MRI) brain were normal. Patient 1 had a normal 46,XX karyotype at 550-band resolution.
Patient 2 is the second child of a 29-year-old-mother without medical report in her family. She has been treated for recurrent depressive episodes with fluotexine for several years. As soon as pregnancy was reported she was given citalopram. The girl is the first child of her 28-year-old-father. Family history was unremarkable. The pregnancy was reported as normal with delivery at term. Birth weight and length were normal. In the first hours of life, she experienced respiratory distress leading to the diagnosis of great vessels transposition. Full surgical correction was done at 2 weeks. At 2 months of age she suffered of an episode of convulsion without fever. The EEG was reported as abnormal (data not available). A brain MRI performed at the age of 10 months showed a thin corpus callosum. Autistic behaviours were reported early on, with poor eye contact contrasting with normal VEP. Smiling was first reported at the age of 4 months and the oral language was completely absent at the age of 28 months. At the age of 2 years, poor social relations were reported with stereotypies of the hands. Sleep was irregular with multiple nocturnal wake-up. She poorly reacted to noises and sounds without anomalies of auditory evoked responses. Motor milestones were also delayed: she sat at the age of 21 months and walked at 3 years 3 months. Craniofacial dysmorphism included trigonocephaly, temporal narrowing, mild hypertelorism, and epicanthic folds (Figure 1b). Metabolic evaluation was reported as normal at the age of 2 years. High-resolution cytogenetic evaluation was normal (600 band level). FISH studies using different probes, that is, 17p11 (Smith–Magenis syndrome), 22q11 (DiGeorge syndrome), 4p16 (Wolf–Hirschhorn syndrome) were normal. Methylation pattern study at 15q11-13 was also normal just as the molecular evaluation at the MECP2 locus.
Molecular cytogenetic investigations
Blood samples were obtained from patients and controls according to our institutional ethical committee and after appropriate informed consent. Among the two patients with submicroscopic 20qter deletion, subtelomeric regions were screened in patient 1 with the Totel Vysion FISH Probe panel (Vysis Inc., Downer's Groves, IL, USA) as instructed by the manufacturer. Array CGH analysis was performed in patient 2 using 1 Mb resolution human bacterial artificial chromosome (BAC) microarray, according to the manufacturer's recommendations (Spectral Genomics, Inc., Houston, TX, USA). Patient and reference (normal female) blood sample genomic DNAs were each split in two samples for differential labelling with Cy3 and Cy5 labeled nucleotides (Amersham Biosciences, Orsay, France) according to standard DNA extraction and random priming protocols. The patient Cy3-labeled DNA was mixed with the Cy5-labeled reference DNA and the patient Cy5-labeled DNA was mixed with the Cy3-labeled reference DNA. Each mix was denatured, and applied to the slides. Slides were hybridised for 48 h, washed in 50% formamide, 2XSSC solutions at 45°C, and scanned on a GenePix 4000B Axon scanner. Images were analysed with SpectralWare software provided by spectral Genomics. Validation and characterisation of the observed chromosome imbalance was achieved by FISH analysis with probes obtained from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk) and selected according to their position on chromosome 20q13.33 according to publicly available genome resources (NCBI Map Viewer: http://www.ncbi.nlm.nih.gov; Santa Cruz Human Genome Browser: http://genome.ucsc.edu). BAC DNA was labelled with biotin by nick translation. The labelled probes were visualized with fluorescein isothiocyanate–avidin (Vector Laboratories, Burlingame, CA, USA).
Results
Patient 1: subtelomeric screening with the Totel Vysion FISH Probe panel revealed an isolated subtelomeric 20q deletion: 46, XX. ish del(20)(qter)(20QTEL14-)dn (not shown). Parental 20qter FISH studies revealed a normal pattern in both parents.
Patient 2: an array CGH analysis was performed with 1 Mb resolution human DNA microarrays as described previously.2 Four probes identified as lost were RP4-583P15, AL11806.27, RP1-81F12 and AL137028 (Figure 2). Parental FISH study confirmed de novo 20q deletion.
(a) Chromosome 20 ratio plot with deviations from the expected 1:1 ratio from clone RP4-583P15 to clone AL137028 (circled in black). FISH to metaphase chromosomes of patient 1 (b) and patient 2 (c) using the BAC RP11-261N11 probe containing CHRNA4 and ARFGAP1 genes (arrows) and a 20 centromeric control probe (Vysis) (arrowheads) showed the deletion of one signal of the BAC RP11-261N11 probe in patient 2 (c).
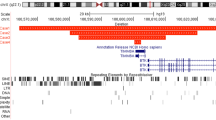
To analyse the chromosome deletions at a higher resolution, BAC clones mapping within and immediately adjacent to the deleted segment were hybridized to metaphase and interphase nuclei from cultured peripheral leucocytes from the patients (Figure 3). Following BAC clones were used, from the centromere to the telomere, RP11-325F17, RP5-885L7, RP11-305P22, RP11-402J8, RP11261-N11, RP11-365O20, RP11-358D14, RP4-583P15, RP5-824A14, RP11-465D20, RP6-1022E24 and RP11-476I15. Patient 1 breakpoint mapped between clone RP11-261N11 and RP11-365O20, whereas Patient 2 breakpoint mapped between clone RP11-305P22 and RP11-402J8. Both patients share a deleted region of approximately 1.33 Mb including 40 confirmed and predicted genes. In these two patients, the deletions differ in 324 kb encompassing five genes, YTDF1, BIRC7, ARFGAP1, COL2A1 and CHRNA4 (Figure 3). The RP-11261N11 probe containing ARFGAP1 and CHRNA4 genes was only deleted in patient 2 (Figure 2).
Map of the 20q13.33 region showing the relative position of Ref Seq genes, BAC clones and genomic variants. BAC clones RP4-583P15, AL118506.27, RP1-81F12 and AL137028 are present on Spectral Genomics microarrays. The position of the deletion breakpoint is indicated in patient 1 (P1bp) and patient 2 (P2bp). The drawing is based on the UCSC map, March 2006 release (http://genome.cse.ucsc.edu/), and the Database of Genomic variants (http://projects.tcag.ca/variation/).
Discussion
To best our knowledge, only three patients with a pure deletion limited to the terminal segment of 20q have been reported in the literature but unfortunately without any accurate size (Table 1). A 9-year-old male had global developmental delay without seizures.3 He sat at 8 months, walked at 15 months, had first words at age 3, and began using short sentences at age 5. At age 7, cognitive testing placed him in the mentally retarded range. On physical examination, he had a frontal upsweep of the hair, up-slanting palpebral fissures, slightly arched eyebrows, bulbous nose, smooth short philtrum and a thin upper lip with a slight Cupid's bow. The deletion was identified by use of subtelomeric probes, but the probes used were not reported. Two other de novo 20qter deletions cases4 were reported using the Vysis Totel Vysion probe panel in a large study population, with only a clinical indication, that is, multiple congenital anomalies in a 11-year-old boy (case 142), and developmental delay and mental retardation in a 7-year-old boy (case 143). In addition, a more complex imbalance was reported in a 7-year-old girl5 carrying a 1.6 Mb deletion of the terminal region of 20q deletion with a 4.7 Mb duplication of the terminal segment of 20p inherited from a familial pericentric insertion of chromosome 20. She suffered generalized tonic seizures, and also had mild ataxia, psychomotor development delay, aggressive and self-injurious behaviour. There was a discrete facial dysmorphism with a large forehead, mild hypertelorism, long prominent nose, short upper lip and dysplastic ears. Finally, a larger terminal deletion (q13qter) was associated with severe seizures, facial dysmorphism and malformation of the extremities.6
The 1.33 Mb deleted region identified in our patients encompasses approximately 40 confirmed and predicted genes including three with known phenotype: SOX18, KCNQ2 and CHRNA4. Different mutations in the voltage-gated potassium channel gene KCNQ2 have been identified in the syndrome benign familial neonatal convulsions, a mild autosomal dominant epilepsy with incomplete penetrance (OMIM 121200). Our two patients have a deletion of the KCNQ2 gene, but only patient 2, at 2 months of age, suffered of an episode of convulsions without fever. Variable expressivity is a common feature of epilepsy caused by ion channel mutations in human patients.7
Patient 2 deletion includes the genes CHRNA4, ARFGAP1 and KCNQ2. Autistic behaviours were reported early on and particularly her sleep was irregular with multiple nocturnal wake-up. Mutations in the α-4 subunit of the neuronal nicotinic acetylcholine receptor gene CHRNA4 have been reported in autosomal-dominant nocturnal frontal lobe epilepsy (ADNFLE; OMIM 6005 513), a syndrome whose clinical diagnosis is often very difficult because of the nocturnal manifestation of symptoms. Mental retardation has been associated with a heterozygous CHRNA4 Ser252Leu mutation in a Korean kindred with ADNFLE.8
The consequences of a decrease in the ADP-ribosylation factor GTPase activating protein 1 gene (ARFGAP1) expression have not been characterized.9
A mouse model is provided by the Stz1 mutation, a spontaneous 300 kb deletion on mouse chromosome 2 that includes Kcnq2, Arfgap1 and Chrna4.10 Mice heterozygous for the Stz1 mutation have increased susceptibility to seizures similar to that seen in Kcnq2 null heterozygotes, whereas Chrna4 null heterozygotes have not been reported to be seizure-sensitive. Interestingly, the 20qter deletion described by Ardalan et al5 included the genes KCNQ2, ARFGAP1and CHRNA4 in a patient with a seizure disorder. Haploinsufficiency for CHRNA4 in addition to loss of KCNQ2and ARFGAP1 may account for the more severe clinical features with seizures. Finally, mutations in the transcription factor gene SOX18 has been shown in three families to underlie recessive and dominant forms of hypotrichosis–lympedema–telangiectasia.11 The absence of these symptoms in our two patients (and in the four patients reported in the literature with pure terminal 20q deletion) suggests that haploinsufficiency for SOX18 may not cause hypotrichosis–lymphedema–telangiectasia, or causes milder disease.
In summary, genotype–phenotype correlations have narrowed the critical interval for the clinical spectrum associated with 20q13.33 deletion. Two clinically distinct phenotypes may be drawn, a mild mental retardation without specific dysmorphism or a more complex and severe phenotype with behaviour problems and seizure disorders, according to the presence or absence of the CHRNA4 and ARFGAP1 genes, respectively. Terminal deletions of 20q seem to be a rare event. Actually, some deletions are simply not looked for in the right population because of the mild mental retardation. However, it also may indicate that the underlying DNA structure in 20q13.33 is less amenable to breakage. A more detailed phenotype description and molecular characterization of further patients with 20q13.33 deletion may help to confirm these data.
References
De Vries BBA, Winter R, Schinzel A, Van Ravenswwaaij-Arts C : Telomeres: a diagnosis at the end of the chromosomes. J Med Genet 2003; 40: 385–398.
Bonnet C, Zix C, Gregoire MJ et al: Characterization of mosaic supernumerary ring chromosomes by array-CGH: segmental aneusomy for proximal 4q in a child with tall stature and obesity. Am J Med Genet 2006; 140A: 233–237.
Roberts AE, Cox GF, Kimonis V, Lamb A, Irons M : Clinical presentation of 13 patients with subtelomeric rearrangements and a review of the literature. Am J Med Genet 2003; 128A: 352–363.
Ravnan JB, Tepperberg JH, Papenhausen P et al: Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet 2003; 43: 478–489.
Ardalan A, Prieur M, Choiset A, Turleau C, Goutieres F, Girard-Orgelet S : Intrachromosomal insertion mimicking a pericentric inversion: molecular cytogenetic characterization of a three break rearrangement of chromosome 20. Am J Med Genet 2005; 138A: 288–293.
Fraisse J, Bertheas MF, Frere F, Lauras B, Rolland MO, Brizard CP : Partial monosomy 20q: a new syndrome. Regional assignement of the adenosine deaminase (ADA) locus on 20q13. Ann Genet 1981; 24: 216–219.
Turnbull J, Lohi H, Meisler MH, Kearney JA, Minassian BA : Sacred disease secrets revealed: the genetics of human epilepsy. Hum Mol Genet 2005; 14: 2491–2500.
Cho YW, Motamedi GK, Laufenberg I et al: A Korean kindred with autosomal dominant nocturnal frontal lobe epilepsy and mental retardation. Arch Neurol 2003; 60: 1625–1632.
Yang JS, Lee SY, Gao M et al: ARFGAP promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol 2002; 159: 69–78.
Yang Y, Beyer BJ, Otto JF et al: Spontaneous deletion of epilepsy gene orthologous in a mutant mouse with a low eletroconvulsive threshold. Hum Mol Genet 2003; 12: 975–984.
Irrthum A, Devriendt K, Chitayat D et al: Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet 2003; 72: 1470–1478.
Acknowledgements
We thank the staff of all contributing French clinical cytogenetics laboratories for their time and effort in submitting data. We also thank to the patients and their family for their cooperation. This study was supported by a regional PHRC-Nancy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Béri-Deixheimer, M., Gregoire, MJ., Toutain, A. et al. Genotype–phenotype correlations to aid in the prognosis of individuals with uncommon 20q13.33 subtelomere deletions: a collaborative study on behalf of the ‘association des Cytogénéticiens de langue Française’. Eur J Hum Genet 15, 446–452 (2007). https://doi.org/10.1038/sj.ejhg.5201784
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201784
Keywords
This article is cited by
-
A novel de novo 20q13.32–q13.33 deletion in a 2-year-old child with poor growth, feeding difficulties and low bone mass
Journal of Human Genetics (2015)