Abstract
Western redcedar (Thuja plicata, Cupressaceae) is a self-fertile conifer with a mixed mating system and significant variation for outcrossing among populations. In this paper, we conducted a fine-scale study of mating system variation to identify correlates of outcrossing in natural populations. We examined variation for outcrossing within and among individual trees, and describe a new method to estimate outcrossing using bulked DNA samples. Bulking (assaying DNA tissues from several individuals simultaneously) increases the experimental power without increasing the experimental effort. We sampled 80 trees from four natural populations in southwestern British Columbia. From each tree, we sampled from up to six crown positions (three heights and inner vs outer branches). From each position, two samples of three seedlings each were bulked before DNA extractions. Using four microsatellite loci, we obtained outcrossing rates for each tree and for each of the six crown positions. We found individual tree selfing rates to increase with tree height in all four populations, but selfing rates did not differ among crown positions. The higher selfing rate of larger trees is probably due to their greater proportional contribution to local pollen clouds. Individual tree outcrossing rates ranged from 22 to 100% and the population outcrossing rates from 66 to 78%. Missed alleles due to bulking and the estimation method used both cause a downward bias in outcrossing rates, so that these estimates are probably lower than the actual outcrossing rates. Nevertheless, the trends we observed are not affected by systematic biases of estimation.
Similar content being viewed by others
Introduction
Western redcedar (Thuja plicata Don ex D. Donn, Cupressaceae) shows quite high selfing for a conifer, with estimates of outcrossing in natural populations ranging from 17 to 100% (weighted mean=71.5%) (O'Connell et al, 2001). The outcrossing rate for most conifer species is above 80% (52 species, mean 83.5%; reviewed in O'Connell, 2003). While western redcedar produces abundant viable selfed seeds (Owens et al, 1990), inbred trees produced by self-fertilization have shown significant reduction in growth rate (ca. 10%) compared to noninbred trees (Russell et al, 2003). A proportion of seeds used for reforestation are from natural populations, thus knowledge of the variation in selfing rates within a tree is important for the collection of seeds with the highest expected outcrossing rate. In addition, knowledge of fine-scale variation of outcrossing rate will enable the understanding of mechanisms promoting selfing, and hence factors affecting the evolution of selfing (Barrett and Eckert, 1990).
The mating system of conifers is dynamic in space and time, mainly being affected by variation of self-pollen availability (Mitton, 1992). Prepollination factors that can affect inbreeding rates in conifers include population density (Farris and Mitton, 1984), crown position (Shaw and Allard, 1982; Omi and Adams, 1986; Chaisurisri et al, 1994), family structure and tree size (Mitton, 1992). In western redcedar, both male and female cones are distributed throughout the crown of trees and the lower branches often reach the ground. Higher selfing is expected in lower branches or in cones closest to the trunk because they are more likely to receive self-pollen from branches higher up in the tree. Likewise, higher branches and branch tips are expected to receive more unrelated pollen and show less selfing (El-Kassaby et al, 1986; Omi and Adams, 1986; Chaisurisri et al, 1994). Higher outcrossing rates in the upper crown vs lower crown have been observed in Sitka spruce (Chaisurisri et al, 1994) and Douglas fir (Shaw and Allard, 1982; Omi and Adams, 1986). In a study of western redcedar seed orchards, selfing rates were found to be higher in lower branches compared to higher branches (K. Ritland, unpublished data). Seed orchards may be unrepresentative of natural populations, however. In another study, differences for outcrossing among natural populations of western redcedar suggested that populations with larger trees had higher selfing rates than populations with smaller and younger trees (O'Connell et al, 2001).
In the past, population outcrossing rates have been estimated with isozyme markers, as in O'Connell et al (2001). More recently, microsatellites have gained popularity for use in plant mating system studies because of their high variability allowing for finer-scale studies. In western redcedar, only one isozyme locus of 21, G6PDH (glucose-6-phosphate dehydrogenase), shows sufficient polymorphism for estimation of outcrossing (Copes, 1981; Yeh, 1988; El-Kassaby et al, 1994). Several highly variable microsatellite loci have been developed for western redcedar, which should allow us to conduct detailed studies of variation of selfing rate (O'Connell and Ritland, 2000). Microsatellites have the statistical power to estimate outcrossing at different crown positions (Pinus densiflora, Pinaceae; Lian et al, 2001), to discriminate between biparental inbreeding and self-fertilization (Caryocar brasiliense, Caryocaraceae; Collevatti et al, 2001), to infer differences of selfing between immature and mature fruits (Shorea leprosula, Dipterocarpaceae; Nagamitsu et al, 2001), and to jointly estimate outcrossing and relatedness among siblings (Zostera marina, Zosteraceae; Reusch, 2000, 2001).
However, compared to the less-informative isozymes, microsatellites are more expensive and labor intensive, often leading to a reduction in sample size compared to isozyme studies. For example, in other microsatellite studies, seeds were sampled from only one maternal tree in Pinus densiflora (874 offspring; Lian et al, 2001), five trees in Shorea leprosula (199 offspring; Nagamitsu et al, 2001) and outcrossing rates were estimated from a single progeny per family in Zostera marina (100 families; Reusch, 2000, 2001). Bulking, or the use of more than one individual in DNA extraction and assay, has been shown to increase the efficiency of gene frequency and heterozygosity estimates (Ritland, 2002). Likewise, for estimating outcrossing, bulking the offspring within a family can significantly increase experimental power for the same effort, provided that alleles can still be detected in bulked samples.
In this study, we test for fine-scale variation of outcrossing rates within natural populations of T. plicata, at the level of individual trees and within individual trees. To increase experimental efficiency, we use a new estimation method based upon the bulking of several individual progeny into one sample. This method greatly increases the power to detect fine-scale variation, as more individuals can effectively be included for the same number of genetic assays.
Materials and methods
Bulking tests
To evaluate the feasibility of bulking DNA (assaying DNA tissues from several individuals simultaneously), we performed three tests to assess whether all alleles could be detected, using samples bulked either before or after DNA extraction. First, we bulked equal proportions of DNA separately extracted from three individuals with known genotypes to test whether all alleles could be detected. Second, we bulked DNA from two individuals in 1:5 DNA ratios to test whether alleles occurring at a low frequency could be detected. Third, we screened samples of one, three or 10 seedlings bulked before DNA extraction, and in this case the individual seedling genotype was not known, but the maternal genotypes were obtained from separately screened haploid megagametophytes.
All individuals were screened at four easily interpretable and robust microsatellite loci (TP1, TP3, TP9, TP11; O'Connell and Ritland, 2000). PCR amplification and detection of alleles on a LI-COR 4200 (Lincoln, Nebraska) were carried out as described in O'Connell and Ritland (2000). To score alleles, we used a program that gives the intensity of each individual band (Odyssey ver. 1.0.55, LI-COR Inc., Lincoln, Nebraska). The band intensity is equal to the sum of the intensity values for all pixels in an area covered by a band (integrated intensity). To test whether alleles were missed in bulked DNA samples compared to unbulked samples, paired t-tests were used, with the number of alleles as units in the tests. Statistical tests were performed using JMP version 3.2.1 (SAS Institute, 1997).
Sample collections
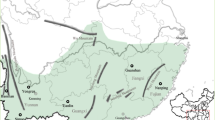
In the autumn of 1999, mature cones were collected from four southwestern British Columbia, Canada, populations: three on eastern Vancouver Island (Paldi, Coombs and Yellow Point) and one on the mainland near Pemberton (O'Connell, 2003). All populations were mixed-species, second-growth stands with western redcedar being a dominant major component co-occurring with Douglas-fir (Pseudostuga menziesii), western hemlock (Tsuga heterophylla) and grand fir (Abies grandis). Sampled trees ranged in height from 4.8 to 36.8 m, and were representative of the height of reproductive individuals in the populations. Cones were collected from up to three different crown heights within a tree: the top of each tree, midway up the tree and from the lowermost branches of the tree (Figure 1). From shorter trees, only two heights (top and bottom) were sampled. At each height, cones were collected from two positions: from the tip of the outer branches and from the hanging inner branches closer to the trunk. In the three island populations, cones were collected from up to six positions on each tree; in the Pemberton population, seeds were collected from only two positions (1 and 5; Figure 1).
Seeds were mechanically extracted from the cones, stored at 4°C until they were germinated in Petri dishes on moist filter paper at room temperature (O'Connell et al, 2001). A few days after germination, the haploid megagametophytes and diploid seedlings were separated and placed in 1.5 ml microtubes. Bulks of 10 megagametophytes were used to obtain maternal genotypes, and bulks of three seedlings to obtain estimates of outcrossing. Samples were kept at −20°C until DNA extraction. Each sample was ground and DNA extractions were carried out using a modified CTAB method in the microtubes (Doyle and Doyle, 1987).
DNA assay of bulks
To test for differences in outcrossing among crown positions, two collections of three bulked seedlings each were screened from each position. Seedlings were screened at four loci: TP1, TP3, TP9 and TP11. Megagametophytes were also scored at three additional loci as part of another study (O'Connell and Ritland, 2004). This information was used to calculate genetic diversity and inbreeding coefficients for each population. The total number of seedlings sampled per tree ranged between 12 (two positions) and 36 (six positions). To ensure that bands were uniformly scored, on each gel we ran an allelic ladder composed of two to three individuals with alleles of known sizes and spanning the range of allele sizes at a locus. Microsatellite alleles typically show additional nonallelic bands of lower intensity and smaller size on gels. These stutter bands are the result of slippage during PCR amplifications. The intensity and size of microsatellites were scored using the Odyssey software. Using the intensity of each band to score alleles helps to differentiate between allelic and nonallelic stutter bands. This extra caution in scoring bands was needed, as many alleles are possible in bulk samples. For example, in a bulk of three seedlings, up to four alleles can exist for a homozygous mother, and up to five alleles for a heterozygous mother.
Estimation of outcrossing from bulked samples
Probabilities of progeny, conditioned upon maternal genotype, are the basic ingredients for the estimation of mating systems. These are functions of the population allele frequencies and the outcrossing rate. Table 1 gives these probabilities for cases of (1) a single progeny, (2) two progeny and (3) three progeny. Ritland (2002) presented a model for estimating gene frequencies and heterozygosities using bulked samples. In that paper, a general probability was given using a ‘mask’ function, but that notation cannot be easily applied to these mating system probabilities; instead, the direct probabilities are given for the simpler cases of bulks of 1, 2 or 3.
The probabilities of single progeny are used in the classic method of estimating outcrossing, while the two- and three-progeny probabilities represent the cases where two and three individuals are bulked, respectively. Using these probabilities, the procedure for estimating outcrossing then follows the usual procedures, described in Ritland (1983). A computer program was written in FORTRAN95 by K Ritland for estimating outcrossing rate for the case of bulks of size three. Maternal parentage was assumed known, and the bootstrap method was used to ascertain errors of estimates. To check the accuracy of the estimation program, a second program was written to simulate data with known parameters.
Results
Allele detection
In samples of two and three seedlings with known genotypes bulked after DNA extraction, all alleles were detected at each of the four loci in both the 1:1:1 and the 1:5 DNA ratios (results not shown). Tests performed with individuals of known genotypes showed that stutter bands were almost exactly half the intensity of the previous band, and the intensity of overlapping bands was additive. Using this information, alleles can be disentangled from stutter bands and other alleles in bulked samples (Figure 2). The tests also showed that, for bulked samples, the number of copies of a particular allele could not be accurately inferred. This is probably because of allele competition during PCR reactions. Bands from larger size alleles are usually less intense than for shorter alleles.
The total number of alleles detected for the same number of seedlings was greater when samples were not bulked (Table 2). Maternal alleles and common alleles in the population were detected in both bulked and nonbulked samples. Low frequency alleles, however, were probably missed in bulked samples. Bulks of three seedlings were chosen for outcrossing rate estimation because individual alleles were easily identified, yet bulking still provided the advantage of reducing the number of samples of DNA to extract and score.
Genetic diversity
Microsatellites were scored in a total of 2019 seedlings (673 bulked assays) from 80 families in four populations (20 families/pop). Expected heterozygosities and inbreeding coefficients for each population based on seven loci were obtained from the megagametophytes (Table 3). Three of the populations had inbreeding coefficients that were significantly greater than zero. The four microsatellite loci used to score seedlings showed a large amount of polymorphism within sampled populations. The number of maternal alleles per population ranged from five to seven at locus TP11, and from 14 to 16 at locus TP9. At each locus, almost twice the number of alleles were detected in the seedlings compared to the maternal plants.
Outcrossing rates
The estimates for outcrossing rate at each crown position were estimated over all trees in a population. Outcrossing rates did not significantly differ among tree position in any population (Table 4). In the three populations where cones were collected separately from inner and outer crowns, outcrossing estimates appeared higher in inner branches compared to outer branches (six of nine positions), but this difference was not statistically significant (paired t-test: 1.57, P=0.077). In contrast, there was a significant correlation between tree height and selfing rates. Over all populations, individual tree outcrossing rates decreased significantly with tree height (r2=0.155, N=73, P=0.0006; Figure 3). An analysis of covariance showed that there was no difference among populations in the mean tree outcrossing rate (F=0.35; df=3; P=0.79) or for the slope of the regression of outcrossing rate on tree height (F=0.19, df=3, P=0.90; Figure 3). Individual tree outcrossing rates range from 22 to 100% and the mean outcrossing rate over all trees in a population ranged from 66 to 78% (Table 5).
The simulation program, written to evaluate the statistical properties of the bulk method, did find some downwards bias for outcrossing, but this bias was not more than 5% of the true value of outcrossing rate, for the specific bulk size of three. This bias is expected with the nonlinearity of the model; in a bulk of three, terms of order t3 appear, for t the outcrossing rate.
Discussion
Within tree variation in outcrossing
No significant difference in outcrossing was detected among crown positions. Actually, all trends observed were in the opposite direction of what was expected. Outcrossing rates were higher in the lower branches vs the higher branches, and higher in inner vs outer branches. Unlike natural populations, trees in a previous study in a western redcedar seed orchard showed a significant decrease in selfing in higher branches compared to lower branches (K Ritland, unpublished data). Higher outcrossing in upper crowns vs lower crowns was also found in a Sitka spruce seed orchard (Picea sitchensis; Chaisurisri et al, 1994) and in Douglas-fir seed orchards and natural populations (Pseudotsuga menziesii, Shaw and Allard, 1982; Omi and Adams, 1986). We expect that the variation in the number of inbred seedlings among different positions within a tree is due mainly to variation in the amount of self-pollen received. Factors in seed orchards that differ from natural populations and that could enhance outcrossing include higher tree density, top pruning and decreased family structure.
Among tree variation in outcrossing
Individual tree outcrossing rates varied widely within all populations ranging between 22 and 100% (Table 5; Figure 3). Among tree differences in rates of selfing can be a result of both among tree variation in self-pollination and self-fertility (O'Connell, 2003). In T. plicata, outcrossing rates decreased with increasing tree height in all four populations. Larger trees were probably more likely to self-fertilize than shorter trees because of their greater proportional contribution to the local pollen cloud. Conversely, smaller trees could receive greater amounts of unrelated pollen from surrounding taller trees. All four populations were similar in terms of tree height structure (Figure 3; Table 5). Phenological differences that occur with tree size can also potentially affect outcrossing. In Douglas-fir, shorter trees have a higher proportion of male cones, and larger trees have more female cones so that outcrossing rate is expected to increase with tree height (Mitton, 1992). The change in outcrossing rates expected in Douglas-Fir is opposite to the trend observed in western redcedar. No data are available on whether there is a change in cone sex ratio with tree size in western redcedar, however, no obvious change has been observed. Furthermore, this species produces copious amounts of both male and female cones throughout the crown once sexual maturity has been reached. Alternatively, if selfing is prevented by a self-incompatibility mechanism, a breakdown in this mechanism with tree age could explain the increase in selfing in larger trees. Although there is no strong evidence for this scenario, it would be an interesting question to pursue.
Population outcrossing rates
Unlike a previous study conducted with isozymes (O'Connell et al, 2001) in six different western redcedar populations, there was no difference in outcrossing rates among populations. In the current study, outcrossing rates were not significantly different among populations and occurred within a narrow range (t=66.2–77.8%). In contrast to the isozyme study, populations sampled in this study were more homogeneous in height structure. If selfing rates are constant over time, the level of inbreeding should be reflected in the inbreeding coefficient. There was no relationship between mean population outcrossing rates and inbreeding coefficients or genetic diversity in this study.
Bulking samples
Our bulking tests showed that most alleles are detected in bulked assays, however, some rare alleles are probably missed. Significantly fewer alleles were detected in bulked samples compared to individually genotyped samples. Estimates of outcrossing obtained using bulked DNA samples are expected to be lower than when using unbulked samples for two reasons. First, alleles are more likely to be missed in bulked samples due to allele competition during PCR and overlapping stutter bands on gels. Second, simulations showed a 5% downward bias in outcrossing estimates of bulked samples. Correspondingly, in the western redcedar orchard study, average outcrossing rates estimated using microsatellites with nonbulked samples were about 5% higher than those observed in this study (K. Ritland, unpublished data). Likewise, in the isozyme study, outcrossing estimates were above 77% for all populations but one (5/6) (O'Connell et al, 2001). However, the comparison of trends in ecological factors affecting outcrossing rates is little affected by systematic statistical bias.
References
Barrett SCH, Eckert CG (1990). Variation and evolution of mating systems in seed plants. In: Kawano S (ed) Biological Approaches and Evolutionary Trends in Plants, Academic Press: London. pp 229–254.
Chaisurisri K, Mitton JB, El-Kassaby YA (1994). Variation in the mating system of Sitka spruce (Picea sitchensis): evidence for partial assortative mating. Am J Bot 81: 1410–1415.
Collevatti RG, Grattapaglia D, Hay JD (2001). High resolution microsatellite based analysis of the mating system allows the detection of significant biparental inbreeding in Caryocar brasiliense, an endangered tropical tree species. Heredity 86: 60–67.
Copes DL (1981). Isoenzyme uniformity in western red cedar seedlings from Oregon and Washington. Can J For Res 11: 451–453.
Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15.
El-Kassaby YA, Parkinson J, Devitt WJB (1986). The effect of crown segment on the mating system in a Douglas-Fir (Pseudotsuga menziesii (Mirb.) Franco) seed orchard. Silvae Genet 35: 149–155.
El-Kassaby YA, Russell J, Ritland K (1994). Mixed mating in an experimental population of western red cedar, Thuja plicata. J Hered 85: 227–231.
Farris MA, Mitton JB (1984). Population density, outcrossing rate, and heterozygote superiority in ponderosa pine. Evolution 38: 1151–1154.
Lian C, Miwa M, Hegetsu T (2001). Outcrossing and paternity analysis of Pinus densiflora (Japanese red pine) by microsatellite polymorphism. Heredity 87: 88–98.
Mitton JB (1992). The dynamic mating systems of conifers. New For 6: 197–216.
Nagamitsu T, Ichikawa S, Ozawa M, Shimamura R, Kachi N, Tsumura Y, Muhammad N (2001). Microsatellite analysis of the breeding system and seed dispersal in Shorea leprosula (Dipterocarpaceae). Int J Plant Sci 162: 155–159.
O'Connell LM (2003). The evolution of inbreeding in western redcedar (Thuja plicata: Cupressaceae). PhD Thesis, University of British Columbia.
O'Connell LM, Ritland CE (2000). Characterization of microsatellite loci in western redcedar (Thuja plicata). Mol Ecol 9: 1920–1922.
O'Connell LM, Ritland K (2004). Somatic mutations at microsatellite loci in western redcedar (Thuja plicata: Cupressaceae). J Hered 95: 172–176.
O'Connell LM, Viard F, Russell J, Ritland K (2001). The mating system in natural populations of western redcedar (Thuja plicata). Can J Bot 79: 753–756.
Omi SK, Adams WT (1986). Variation in seed set and proportions of outcrossed progeny with clones, crown position, and top pruning in a Douglas-fir seed orchard. Can J For Res 16: 502–507.
Owens JN, Colangeli AM, Morris SJ (1990). The effect of self-, cross-, and no pollination on ovule, embryo, seed, and cone development in western red cedar (Thuja plicata). Can J For Res 20: 66–75.
Reusch TBH (2000). Pollination in the marine realm: microsatellites reveal high outcrossing rates and multiple paternity in eelgrass Zostera marina. Heredity 85: 459–464.
Reusch TBH (2001). Fitness-consequences of geitonogamous selfing in a clonal marine angiosperm (Zostera marina). J Evol Biol 14: 129–138.
Ritland K (1983). Estimation of mating systems. In: Tanksley SD, Orton TJ (eds) Isozyme in Plant Genetics and Breeding, Part A, Elsevier Scientific Publications: Amsterdam. pp 289–302.
Ritland K (2002). Estimation of gene frequency and heterozygosity from pooled samples. Mol Ecol Notes 2: 370–372.
Russell JH, Burdon RD, Yanchuk AD (2003). Inbreeding depression and variance structures for height and adaptation in self- and outcross Thuja plicata families in varying environments. For Genet 10: 171–184.
SAS Institute (1997). JMP user's guide version 3.2.1, SAS Institute Inc.: Cary, North Carolina.
Shaw DV, Allard RW (1982). Estimation of outcrossing rates in Douglas-fir using isozyme markers. Theor Appl Genet 62: 113–120.
Yeh FC (1988). Isozyme variation of Thuja plicata (Cupressaceae) in British Columbia. Biochem Syst Ecol 16: 373–377.
Acknowledgements
The assistance of D Pigott, J Ogg, ‘Zed’ (cone collections), C Ferguson (seed extractions), and M Brown (tree heights) is greatly appreciated. This research was funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) research grant to KR and NSERC and Izaak Walton Killam Predoctoral Fellowships to LMO.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
O'Connell, L., Russell, J. & Ritland, K. Fine-scale estimation of outcrossing in western redcedar with microsatellite assay of bulked DNA. Heredity 93, 443–449 (2004). https://doi.org/10.1038/sj.hdy.6800521
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800521
Keywords
This article is cited by
-
Estimation of Outcrossing Rates in Biodiesel Species Pongamia pinnata Based on AFLP and Microsatellite Markers
National Academy Science Letters (2017)
-
Randomized, replicated, staggered clonal-row (R2SCR) seed orchard design
Tree Genetics & Genomes (2014)
-
Temporal and spatial variation of pollen yield in natural populations of Pinus roxburghii
Forestry Studies in China (2012)
-
Variation of selfing rate and inbreeding depression among individuals and across generations within an admixed Cedrus population
Heredity (2011)
-
Mating systems in tropical forages: Stylosanthes capitata Vog. and Stylosanthes guianensis (Aubl.) Sw.
Euphytica (2011)