Abstract
We genotyped 45 biallelic markers and 11 STR systems on the Y chromosome in 201 male Somalis. In addition, 65 sub-Saharan Western Africans, 59 Turks and 64 Iraqis were typed for the biallelic Y chromosome markers. In Somalis, 14 Y chromosome haplogroups were identified including E3b1 (77.6%) and K2 (10.4%). The haplogroup E3b1 with the rare DYS19-11 allele (also called the E3b1 cluster γ) was found in 75.1% of male Somalis, and 70.6% of Somali Y chromosomes were E3b1, DYS19-11, DYS392-12, DYS437-14, DYS438-11 and DYS393-13. The haplotype diversity of eight Y-STRs (‘minimal haplotype’) was 0.9575 compared to an average of 0.9974 and 0.9996 in European and Asian populations. In sub-Saharan Western Africans, only four haplogroups were identified. The West African clade E3a was found in 89.2% of the samples and the haplogroup E3b1 was not observed. In Turks, 12 haplogroups were found including J2*(xJ2f2) (27.1%), R1b3*(xR1b3d, R1b3f) (20.3%), E3b3 and R1a1*(xR1a1b) (both 11.9%). In Iraqis, 12 haplogroups were identified including J2*(xJ2f2) (29.7%) and J*(xJ2) (26.6%). The data suggest that the male Somali population is a branch of the East African population – closely related to the Oromos in Ethiopia and North Kenya – with predominant E3b1 cluster γ lineages that were introduced into the Somali population 4000–5000 years ago, and that the Somali male population has approximately 15% Y chromosomes from Eurasia and approximately 5% from sub-Saharan Africa.
Similar content being viewed by others
Introduction
East Africans are more related to Eurasians than to other African populations.1, 2, 3 Investigations of Y chromosome markers have shown that the East African populations were not significantly affected by the east bound Bantu expansion that took place approximately 3500 years ago, while a significant contact to Arab and Middle East populations can be deduced from the present distribution of the Y chromosomes in these areas.4, 5 The Y chromosome haplogroup E3a is found at high frequencies in the sub-Saharan, Bantu-speaking populations but at low frequencies in East Africa, while Eurasian haplogroups like J and K are found at various frequencies in East Africa.3, 4, 6, 7, 8, 9 However, the majority of Y chromosomes found in populations in Egypt, Sudan, Ethiopia and Oromos in Somalia and North Kenya (Boranas) belong to haplogroup E3b1 defined by the Y chromosome marker M78.9, 10 A special branch of E3b1, cluster γ, which was defined by the presence of the otherwise rare Y STR allele 11 in DYS19, was observed in high frequencies in small samples of male Boranas (Oromos) in North Kenya, Ethiopian Oromos and Somali males, while the E3b1 cluster γ was found in low frequencies in non-Oromos from Ethiopia, Bantus from Kenya, North Egyptians10 and was almost absent in populations outside the Horn of Africa. Other clusters of haplogroup E3b1 (α, β and δ) that are found in European, Arab, North and East African populations were not found in Oromos from North Kenya (Boranas) or Ethiopia, and found in only one of 23 Somali males.10
We typed a set of 45 biallelic markers and 11 STR systems on the Y chromosome in a large population of male Somali immigrants to Denmark in order to define their Y chromosome lineages in details. In addition, 65 sub-Saharan Western Africans, 59 Turks, and 64 Iraqis were typed for the biallelic Y chromosome markers. The results were compared to those obtained in other relevant populations.
Material and methods
Samples and DNA purification
A total of 389 DNA samples from unrelated males (the numbers of individuals are given in parentheses) from Turkey (59), Iraq (64), Somalia (201) (all immigrants to Denmark) and 65 sub-Saharan Western Africans from Mali (38), Ghana (16), Mauritania (three), Guinea Conakry (two), Liberia (two), Cote d'Ivoire (one), Guinea – Bissau (one), Senegal (one) and Cameroon (one) (all immigrants to the Canary Islands) were typed in duplicates for 45 biallelic Y chromosome markers. Blood on FTA cards or Qiagen-purified DNA was used. The protocol was approved by the Danish ethical committee (Ref. KF-01-037/03).
Biallelic marker typing
The PCR amplification, the single base extension (SBE) reaction and the determination of the biallelic markers were performed as described previously.11 The markers P2, M22, M70, M75, M128, M168, M201, M207, M269 and M304 were also typed using singleplex PCR conditions.4, 11, 12 For the marker V6,10 we used the PCR primer sequences: V6F: 5′-CCTATAGAGTCCCTGTCCCTGA-3′, V6R: 5′-CTTGCTGCTGAGTGAGCTTCT-3′ (0.4 μ M of each primer). SBE primers not described by Sanchez et al11 (0.2 μ M of each primer) are given in Table 1.
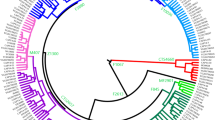
The haplogroup relationships and the nomenclature were established by the Y Chromosome Consortium (YCC).5, 13 The relevant portion of the YCC tree is shown in Figure 1.
Phylogenetic distribution of the 43 Y chromosome haplogroups that can be detected by the 45 biallelic markers. The arrow indicates the ancestral root of the maximal parsimonious YCC tree (2003).5 The major divisions of human Y chromosome diversity are labelled with large, capital letters in bold. On the right is shown the distribution of Y chromosome haplogroups in Somalis and in people from sub-Saharan West Africa, Turkey and Iraq. The relative frequencies in percent are shown in parentheses. aBecause none of our subjects studied belong to the haplogroup E3b1b, defined by the presence of M224,4 we used the haplogroup name E3b1 instead of E3b1*(xE3b1b) in the text.
STR typing
Y STR typing was performed using the PowerPlex® Y System kit (Promega) including DYS391, DYS389I, DYS389II, DYS439, DYS438, DYS437, DYS19, DYS392, DYS393, DYS390 and DYS385. The PCR conditions were as recommended by the manufacturer except that the number of cycles was 10 plus 16. The amplifications were performed in a Perkin–Elmer GeneAmp® PCR System 9700 thermal cycler. A total of 1 μl of PCR product was electrophoretically separated on an ABI 3100 Genetic Analyzer (Applied Biosystems) using performance-optimized polymer 4 (POP4) and Dye Set Z. Analyses of PCR fragments were performed using GeneScan 3.7 and Genotyper 3.7 NT (Applied Biosystems). The alleles were assigned by comparison to allelic ladders using Genotyper macros supplied with the PowerPlex® Y System. All 201 Somali males and the Turkish and Iraqi males previously assigned to belong to the haplogroup E3b1 were typed in duplicate. The nomenclature for DYS389II reflects the total number of repeats minus the number of repeats of DYS389I.14
As suggested by de Knijff,15 Y chromosomes identified by STRs are designated ‘haplotypes’. Y chromosomes that are defined only by biallelic markers are called ‘haplogroups or clades’ and the combination of biallelic markers and Y-STRs are called ‘lineages’.
Typing of 15 autosomal STRs was performed using the AmpFlSTR Identifiler PCR Amplification Kit (Applied Biosystems) according to the instructions of the manufacturer. A total of 198 of the 201 Somali males were typed in duplicate. The last three Somali males were not typed due to technical problems.
Comparative data analyses
In order to compare the proportions of Y-STR haplotypes in Somalis with those in other populations, we searched the worldwide Y-STR Haplotype Reference Database (http://www.yhrd.org/index.html). In addition, we compared the Somali Y STR data with results in African and Anatolian populations available in the literature. Phylogenetic relationships between Somali STR haplotypes within haplogroups were reconstructed in a median-joining network16 using the programme Network 4.1.0.0 (http://www.fluxus-technology.com). A haplogroup-specific weight proportional to the reciprocal of the microsatellite variance was used. Reduced-median and median-joining (ɛ=0) procedures were applied sequentially.
Statistical analyses
We used GENEPOP version 3.417 to test for Hardy–Weinberg proportions of the alleles of the 15 autosomal STR systems and calculations of the Fis values.18
The software package Arlequin, version 2.00019 (http://lgb.unige.ch/arlequin) was used to determine the molecular variance (AMOVA) at various levels of hierarchical groupings based on haplogroup frequencies20 and the mean pair wise differences, which are the mean number of mutational steps observed between all pairs of haplotypes in the sample.
The significance of the variance components and the corresponding Φ statistics (F statistics analogs) was assessed by comparisons of the observed values with the distribution of 10 000 permutations obtained by randomization under the null hypothesis of no population structure. The AMOVA was made considering the subpopulation relative to the total population (ΦST) and the geographical group of a subpopulation relative to the total population (ΦCT).6, 8 Principal component analysis of the haplogroup frequencies was performed using the Multivariate Statistical Package (MVSP) v. 3.1 (http://www.kovcomp.co.uk/mvsp/mvspwbro.html) and presented graphically in two dimensions.
Y chromosome STR data were used to estimate the expansion time using both a model that does not explicitly specify a demography (through a star-like genealogy) and a Bayesian-based coalescence analysis procedure21 assuming a stepwise mutation model. The first approach required identification of the ancestral haplotype within each haplogroup using the ΔA statistics described by Stumpf and Goldstein.22 To estimate the time to the most recent common ancestor (TMRCA), we used the average squared difference (ASD)23 and the averaged effective mutation rate described by Zhivotovsky et al.24 We used 95% confidence intervals (CIs) estimated by Monte Carlo simulations using a coalescent model with exponential growth scaled in units of the TMRCA including the uncertainty of the mutation rate. This is implemented in the programme Ytime described by Behar et al.25 Generation times of 25 and 30 years were assumed. Thus, the 95% CI takes into account the uncertainty in mutation rate, the population growth and (where appropriate) subdivision, but not the generation time. We also calculated the TMRCA using the variance of repeat scores observed (averaged over loci) within a haplogroup.26, 27
Bayesian analysis of trees with internal node generation (BATWING)28 was used to estimate the expansion times of a set of Somali Y chromosomes. The demographic model assumed exponential growth from an initially constant-sized population beginning at time Beta. Priors were chosen to be as uninformative as possible in order to minimize the impact on the results. Thus, we specified a gamma (1, 0.001) distribution as the prior for the growth rate and a gamma (1.1, 0.0001) distribution as the initial population size.12, 29 The prior distribution for the STR mutation rate was specified as a gamma distribution with a mean of 6.9 × 10−4 per locus per 25 or 30 years,24 and a broad, uniform Beta prior (0, 15) distribution was assigned. The estimated time of population expansion, Beta, was expressed as a fraction of the initial population size multiplied by the generation time to generate standard units of time. A total of 20 000 initial rearrangements were discarded, and the posterior distributions were estimated from the subsequent 50 000 rearrangements using the R computer software Version 1.8.1 (http://www.r-project.org/).
The results of microsatellite loci DYS385 were excluded from the statistical comparison analysis except for the network analysis because it was impossible to assign alleles to a specific locus. Estimation of the genetic diversity was measured using Nei's unbiased h statistics.30 The variance of the allele distribution was calculated in EXCEL (Microsoft) for each locus independently and then averaged across the 10 loci.
Although the Horn of Africa is considered a geographic part of sub-Saharan Africa, we have analysed the Somali population separately in order to be able to compare the results with previously published data from other African populations.
Results
Autosomal STR typing
The genotypes of the 15 autosomal STR systems in 198 Somalis were in Hardy–Weinberg equilibrium (χ2=31.37, df=30, P=0.40), and the Fis values ranged from −0.047 to 0.038.
Y chromosome haplogroup variation
We identified a total of 23 Y chromosome haplogroups in 389 males from Somalia, sub-Saharan West Africa, Turkey and Iraq. Figure 1 shows the genealogical relationship of the haplogroups and their frequencies.
In Somali males, 14 haplogroups were identified. The frequency of the clade E3b was 81.1%, including 77.6% of the haplogroup E3b1 defined by the M78 mutation. The Eurasian haplogroup K2 was found in 10.4%, and 3.0% of the Somali Y chromosomes belonged to the major clade J. Only 3.0% of the Somalis had the sub-Saharan African haplogroups A3, B and E3a*(xE3a4). Less than 2.0% of the Somalis belonged to the Northwest African E3b2 lineage. In the present study, no individual belonging to E3b* chromosomes carried the V6 mutation, which identifies a subset of chromosomes assigned to E3b* (E-M35*).10
Among the sub-Saharan Western Africans, only four haplogroups were identified. The West African clade E3a was found in 89.2%. Only one individual carried the major clade E3b (1.5%), and the haplogroup E3b1 was not observed.
In Turks, 12 haplogroups were found. The four haplogroups J2*(xJ2f2) (27.1%), R1b3*(xR1b3d, R1b3f) (20.3%), E3b3 and R1a1*(xR1a1b) (both 11.9%) were the most frequent ones.
In Iraqis, 12 haplogroups were identified. The haplogroup J2*(xJ2f2) was the most frequent one (29.7%) followed by J*(xJ2) (26.6%).
Geographic distribution of the Y chromosome haplogroup E3b1
To examine the relationship between the Y chromosome haplogroups in the Somali, other African and non-African populations, we compared our data with the data from literature (Table 2). The high frequency (77.6%) of haplogroup E3b1 was characteristic of male Somalis. The frequency of E3b1 was significantly lower in Ethiopian Oromos (35.9%), Ethiopian Amharas (22.9%), Egyptians (20.0%), Sudanese (17.5%), Kenyans (15.1%),10 Iraqis (6.3%), Northern Africans (6.1%), Southern Europeans (0.5–5.1%) and sub-Saharan populations (below 1%).
We analysed the variation in the frequencies of E3b1 in 17 geographically defined populations using the AMOVA test.20 A total of 61.2% of the variance (significantly greater than zero, P<10−4) could be attributed to differences within populations. The population data were grouped into (1) sub-Saharan Africans (four populations), (2) North and East Africans (seven populations) and (3) non-Africans (six populations), and the Φ statistics were calculated. High degrees of both inter- and intragroup variabilities (ΦCT=0.31, P<10−4; ΦSC=0.19, P<10−4) were observed. In North and East African populations (ΦST=0.25, P<10−4), the variability was mainly due to the high frequency of the haplogroup E3b1 in the Somalis (77.6%) compared to those in Sudanese (17.5%) and Northern Africans (6.1%). When the Somali population data were removed from the North and Eastern African group, the ΦST value decreased from 0.25 to 0.16.
Figure 2 shows a principal component analysis of the haplogroups. We observed a similar pattern in a neighbour joining, unrooted tree (data not shown). The two principal components accounted for 79.4% of the genetic variance observed mainly due to differences in the frequencies of the clade E and clade BR*(xE) (first component) and the E3a and E3b lineages (second component). The first component separated the non-Africans and the sub-Saharan Western Africans characterized by high frequencies of the clade E3a (Figure 2, axis 1). The North and East Africans were separated from the rest of the sub-Saharan populations (Figure 2, axis 2) mainly by different frequencies of haplogroup E3b1. The position of the Ethiopian Oromos was close to that of the Somalis due to the relatively high frequencies of the haplogroups E3b*(xE3b1) and E3b1. The Ethiopian Amharas and the Egyptian population were positioned between the African and the non-African populations, primarily due to the high frequencies of the clade BR*(xE).
Principal component analysis of the relative frequencies of Y chromosome haplogroups in the populations reported in Table 2. The vectors express the relative weight of each haplogroup in the first and/or second axis. The positive or negative values indicate with which end of the axis the haplogroups are associated. Thus, the first principal component (axis 1) explained 52.8% of the total variance, mainly due to differences in the frequencies in clade E and clade BR*(xE). The second component (axis 2) explained 26.6% of the total variance, mainly due to the differences in the frequencies of the E3a and E3b lineages.
Y chromosome haplotype diversity
The allele and haplotype frequencies of 11 Y-STRs were estimated in the 201 male Somalis. Table 3 shows the Y-STR allele frequencies in all males, in haplogroup E3b1 and in non-E3b1 males. In eight of the 11 Y-STR systems, a predominant allele with a frequency above 0.75 was found. The frequencies of the predominant alleles ranged from 0.90 to 0.99 in E3b1 Y chromosomes.
A comparison between the Y-STR allele frequencies in our and another sample of 104 Somali males typed for the eight common Y-STR loci37 showed that the distributions of Y-STRs were very similar in the two populations (Fst=0.0007; P=0.285).
In Somalis, 96 haplotypes were identified with the 11 Y-STRs. The haplotype diversity was 0.9726±0.005. No microsatellite haplotype was found in more than one haplogroup. The haplotype diversity of the eight Y-STRs (minimal haplotype) was 0.9575±0.007 compared to an average of 0.9974 and 0.9996 in European and Asian populations in the ‘‘Y-STR Haplotype Reference Database’ (http://www.yhrd.org/index.html), 0.9884 in the Mozambican population38 and 0.9838 in the Tunisian population.39
Haplogroup E3b1 with the rare allele 11 of DYS19 (E3b1 cluster γ)10 was found in 75.1% of all Somali males, and 96.8% of the E3b1 Y chromosomes carried DYS19-11. The majority of the E3b1 DYS19-11 Y chromosomes were characterized by DYS392-12, DYS437-14, DYS438-11 (96.2%) and further by DYS393-13 (91.0%).
The average difference in numbers of repeat units between STR haplotypes typed for 10 Y-STRs was 0.7±0.1, and the variance in the allele size distribution was 0.59 repeat units indicating that the Somali Y chromosome haplotypes were very closely related to each other. Figure 3a shows a median joining network analysis of the relatedness of the 64 Y STR haplotypes of the E3b1 haplogroup. The network displayed star-like features, and the four most frequent haplotypes, which accounted for 47% of the entire E3b1 cluster γ lineage, occupied central positions in the network.
Networks of the Y chromosome STR data in haplogroup E3b1. (a) The network was constructed with the reduced-median procedure followed by the median-joining procedure based on data of 11 Y STRs in males belonging to haplogroup E3b1 in the present Somali population sample. The grey and black circles represent the Somali Y chromosomes with 12 and more than 12 repeats in the DYS19 microsatellite, respectively. Unless indicated by a number on the pie, the area of each circle is proportional to the number of individuals with the haplotype. The smallest circle corresponds to one Y chromosome. The lengths of the branches are proportional to the number of mutational steps. (b) The network was constructed with the reduced-median procedure followed by the median-joining procedure by combining our data with previously published data of DYS19, DYS389I, DYS390, DYS391 and DYS392 in individuals belonging to haplogroup E3b112, 40 (personal communication). The broken line delimits the lineages characterized by E3b1, DYS19-11 and DYS392-12 in Somalis and Ethiopians.
The network of the Figure 3b was constructed by combining our data with previously published data for individuals belonging to haplogroup E3b1 (12, 40 and personal communication). It displayed star-like features with a clear geographic structure. The main branch of the E3b1 cluster γ lineages is located on a branch defined by DYS392-12 in the Ethiopian part of the network.
The Somali E3b1 haplotype data were compared with results from Anatolia,12 the only E3b1 data available for the same set of Y-STR loci. The two populations shared only one haplotype (DYS19-13/DYS389I-13/DYS389II-17/DYS390-23/DYS391-10/DYS392-11/DYS393-13/DYS439-12). The E3b1 cluster γ lineages were not found in Turkish or Iraqi males.
In the 21 Somali males belonging to haplogroup K2, the Y-STR haplotypes were organized with a common ancestor into three branches with several mutation steps between the haplotypes (data not shown).
Estimates of expansion time and population size
Table 4 presents estimates of ages and expansion times of the Somali E3b1 cluster γ and the K2 lineages based on Y-STR data using different estimation procedures. By defining the ancestral haplotype as that with the modal allele for each STR system and calculating the average squared distance41 as well as the variance between this haplotype and other variants, we estimated the time back to the most recent common ancestor (TMRCA). In Somalis, the TMRCA was estimated to be 4000–5000 years for the haplogroup E3b1 cluster γ and 2100–2200 years for the haplogroup K2 assuming a generation time of 25 years. Calculations based on a Bayesian coalescence approach (BATWING expansion time) indicated that the growth of the E3b1 cluster γ in the Somali population started 1200 years ago (Table 4) with an initial population size of 1037 individuals. A similar analysis of haplogroup K2 resulted in a calculated expansion time of approximately 3300 years in a small male population of 109 individuals. The results did not change significantly when different prior probability distributions were applied (data not shown).
Discussion
The present study demonstrates that male Somalis has the highest frequency of the haplogroup E3b1 (77.6%) observed in any population studied until now. The great majority of the Somali E3b1 Y chromosomes studied (96.8%) carried the otherwise rare allele 11 of the DYS19 STR locus and, thus, belonged to the cluster γ defined by Cruciani et al.10 The E3b1 cluster γ was previously reported in five of seven (71.4%) male Boranas (Oromos) from North Kenya, in 52.2% of Somali males, and in 32.0% Ethiopian males.10 The majority of the E3b1 Y chromosomes (91.0%) were further characterized by the DYS19-11, DYS392-12, DYS437-14, DYS438-11, and DYS393-13 alleles. The Eurasian clade FR had a frequency of 15.4% and the typical sub-Saharan haplogroups A, B, E3a*(xE3a4), E2 and E3b* were found in only 5% of Somali males.
The network of the E3b1 lineages in the present Somali population sample (Figure 3a) displayed star-like features and we observed a low Y STR haplotype diversity and a very limited spread in the sizes of the STR alleles (Table 3), suggesting a coherent, common, recent ancestry. The network of the E3b1 lineages of previously published data of East African populations and our data (Figure 3b) demonstrate that the E3b1 cluster γ lineages of the present Somali population sample are part of the East African E3b1 lineages. E3b1 cluster γ lineages were observed in low frequencies in Bantus from Kenya, North Egypt, Morocco and Niger10 (Figure 4). In the present study, haplogroup E3b1 was found in 6.3% of Iraqis and none of them belonged to cluster γ. Only 11 subjects with a DYS19-11/DYS392-12 pattern were reported outside the Horn of Africa in 26 654 subjects analysed in a worldwide set of 236 populations by November 2004 (http://www.yhrd.org/index.html). Taken together, the data suggest that the E3b1 cluster γ DYS392-12 lineage was expanded in the Somali population.
Map of African areas where E3b1 cluster γ has been observed (the numbers of individuals are given in parentheses).10 (1) Moroccan Arabs (54), (2) Northern Egyptians (21), (3) Ethiopian Jews (22), (4) Ethiopian Amharas (34), (5) Ethiopian Wolaytas (12), (6) Mixed Ethiopians (12), (7) Ethiopian Oromos (25), (8) Somalia (224 including our Somali data), (9) Boranas (Oromos) from Kenya (seven), (10) Bantus from Kenya (28), (11) Tuaregs from Niger (22). The haplogroups or remaining paragroups are represented by different fill patterns. Lineages excluded from a haplogroup are listed within parentheses after the name of the haplogroup. The distribution of the Cushitic language in East Africa is shown in grey.
Cruciani et al10 suggested that the E3b1 cluster γ lineages originated in East Africa and estimated that the TMRCA was approximately 9600 years. We estimated that the E3b1 cluster γ DYS392-12 lineages of the present Somali population sample originated 4000–5000 years ago, and that the expansion of the E3b1 cluster γ DYS392-12 lineages in these Somalis involved a relatively small number of Y chromosomes (around 1000 males).
The time of the eastbound Bantu expansion was estimated to be 3400±1100 years ago.24 Bantu populations have high frequencies of E3a haplogroups.4 We have observed only a few individuals with the E3a haplogroup in our Somali population, thus, supporting the view that the Bantu migration did not reach Somalia.42 It has been suggested that a barrier against gene flow exist in the region.43 The barrier seems to be the Cushitic languages and cultures to which Somalis belongs. The Cushitic languages belong to the Afro-Asiatic languages that are spoken in Northern and Eastern Africa. The Cushitic languages and cultures are mainly found in the Somalis and the Oromos, one of the two main groups inhabiting Ethiopia.44, 45, 46. The Somali and Oromo languages have a high degree of similarity and the two populations share many cultural characteristics. The Somali and Oromo people live in clans with special patterns of marriage and the Somali and Oromo people have complex, interwoven pedigrees.44, 45
The very high frequency of the E3b1 cluster γ in our Somali population sample could be due to ascertainment bias or special clan or family relationships in the present sample of Somali immigrants to Denmark. No reliable information on geographic origin or clan relationship in the present Somali population sample was available. However, the genotypes of the autosomal STR systems were in Hardy–Weinberg equilibrium, indicating random mating in at least the last generation, and the distribution of Y-STR haplotypes in our Somali population was similar to that in a sample of Somali immigrants to Norway,37 indicating that these two population samples came from a larger, homogenous population of Somalis.
The haplogroup K2 was found in 10.4% of Somali males. Haplogroup K2 was suggested to have arisen in Eurasia.4, 9 K2 has a patchy distribution in Cameroon (18.0%), Egypt (8.2%), Ethiopia (4.8%), Tanzania (3.8%) and Morocco (3.6%), probably due to back migration.3, 7, 8, 9 Luis et al9 estimated an expansion time of 13.7–17.5 ky for the K2 lineages in Egypt. The BATWING expansion time estimated for K2 in our Somali population (3.3 ky) is consistent with an African southward dissemination of the K2 haplogroup. The observation of two Somali males with the M17 mutation (haplogroup R1a1*(xR1a1b)) may indicate a recent gene flow by migration from Eurasia.47, 48 A possible explanation is offered by the fact that from the 7th century onward, immigrant Muslim Arabs and Persians established trading posts along the Somali cost51, although also British, French and Italian people were present in Somalia in the region in the 19th and 20th century.
The distribution of the haplogroups J2*(xJ2f2) (0.5%) and J*(xJ2) (2.5%) in Somalis support the recent gene flow hypothesis. Haplogroup J*(xJ2) was probably spread by the Arab people.40 The ratio between the haplogroups J2/J*(xJ2) may be an indicator of the genetic components from populations like (1) Balkans, Turks, Georgians and Muslim Kurds and (2) Bedouin and Palestinian Arabs, respectively.40, 52 The ratio was 0.26 in the Oman population.9 The J2/J*(xJ2) ratio of 0.2 in the present Somali sample suggest a predominant gene flow of Arab Y chromosomes.
In conclusion, the data suggest that the male Somali population is a branch of the East African population – closely related to the Oromos in Ethiopia and North Kenya (Boranas) – with predominant E3b1 cluster γ DYS392-12 lineages that probably were introduced into the Somali population 4000–5000 years ago, approximately 15% Y chromosomes from Eurasia and approximately 5% from sub-Saharan Africa. Work is in progress in order to study closely related populations with new informative markers to obtain a better understanding of the E3b1 lineages settlement process in East Africa.
References
Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS : Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 1998; 62: 420–434.
Tishkoff SA, Williams SM : Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 2002; 3: 611–621.
Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA : Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 2002; 70: 265–268.
Underhill PA, Passarino G, Lin AA et al: The phylogeography of Y-chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 2001; 65: 43–62.
Jobling MA, Tyler-Smith C : The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003; 4: 598–612.
Scozzari R, Cruciani F, Santolamazza P et al: Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 1999; 65: 829–846.
Underhill PA, Shen P, Lin AA et al: Y-chromosome sequence variation and the history of human populations. Nat Genet 2000; 26: 358–361.
Cruciani F, Santolamazza P, Shen P et al: A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 2002; 70: 1197–1214.
Luis JR, Rowold DJ, Regueiro M et al: The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 2004; 74: 532–544.
Cruciani F, La Fratta R, Santolamazza P et al: Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 2004; 74: 1014–1022.
Sanchez JJ, Børsting C, Hallenberg C, Buchard A, Hernandez A, Morling N : Multiplex PCR and minisequencing of SNPs – a model with 35 Y chromosome SNPs. Forensic Sci Int 2003; 137: 74–84.
Cinnioǧlu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Y Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Rolf B, Meyer E, Brinkmann B, de Knijff P : Polymorphism at the tetranucleotide repeat locus DYS389 in 10 populations reveals strong geographic clustering. Eur J Hum Genet 1998; 6: 583–588.
de Knijff P : Messages through bottlenecks: on the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet 2000; 67: 1055–1061.
Bandelt HJ, Forster P, Rohl A : Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48.
Raymond M, Rousset F : GENEPOP (version 1.2): populations genetics software for exact tests and ecumenicism. J Hered 1995; 86: 248–249.
Robertson A, Hill WG : Deviations from Hardy–Weinberg proportions: sampling variances and use in estimation of inbreeding coefficients. Genetics 1984; 107: 703–718.
Schneider S, Kueffer J-M, Roessli D, Excoffier L : Arlequin: A Software for Population Genetic Data Analysis, 2nd edn. University of Geneva, Switzerland: Genetics and Biometry Laboratory, 2000.
Excoffier L, Smouse PE, Quattro JM : Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992; 131: 479–491.
Zhivotovsky LA : Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 2001; 18: 700–709.
Stumpf MP, Goldstein DB : Genealogical and evolutionary inference with the human Y chromosome. Science 2001; 291: 1738–1742.
Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB : Origins of Old Testament priests. Nature 1998; 394: 138–140.
Zhivotovsky LA, Underhill PA, Cinnioǧlu C et al: The effective mutation rate at Y-chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779.
Kittles RA, Perola M, Peltonen L et al: Dual origins of Finns revealed by Y-chromosome haplotype variation. Am J Hum Genet 1998; 62: 1171–1179.
Ruiz-Linares A, Ortiz-Barrientos D, Figueroa M et al: Microsatellites provide evidence for Y-chromosome diversity among the founders of the New World. Proc Natl Acad Sci USA 1999; 96: 6312–6317.
Wilson IJ, Balding DJ : Genealogical inference from microsatellite data. Genetics 1998; 150: 499–510.
Weale ME, Weiss DA, Jager RF, Bradman N, Thomas MG : Y-chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol 2002; 19: 1008–1021.
Nei M : Molecular Evolutionary Genetics. New York: Columbia University Press, 1987.
Moran CN, Scott RA, Adams SM et al: Y chromosome haplogroups of elite Ethiopian endurance runners. Hum Genet 2004; 115: 492–497.
Arredi B, Poloni ES, Paracchini S : Predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet 2004; 75: 338–345.
Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J : High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 2001; 68: 1019–1029.
Flores C, Maca-Meyer N, Pérez JA, González AM, Larruga JM, Cabrera VM : A predominant European ancestry of paternal lineages from Canary Islanders. Ann Hum Genet 2003; 67: 138–152.
Maca-Meyer N, Sánchez-Velasco P, Flores C et al: Y-chromosome and mitochondrial DNA characterization of Pasiegos, a human isolate from Cantabria (Spain). Ann Hum Genet 2003; 67: 329–339.
Francalacci P, Morelli L, Underhill PA et al: Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol 2003; 12: 270–279.
Stenersen M, Perchla D, Søvik E, Flønes AG, Dupuy BM : Kurdish (Iraq) and Somalian population data for 15 autosomal and 9 Y-chromosomal STR loci. Int Congr Ser 2004; 1261: 185–187.
Alves C, Gusmao L, Barbosa J, Amorim A : Evaluating the informative power of Y-STRs: a comparative study using European and new African haplotype data. Forensic Sci Int 2003; 134: 126–133.
Brandt-Casadevall C, Ben Dhiab M, Taroni F et al: Tunisian population data on 10 Y-chromosomal loci. Forensic Sci Int 2003; 135: 247–250.
Semino O, Magri C, Benuzzi G et al: Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW : Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci USA 1995; 92: 6723–6727.
Newman JL : The Peopling of Africa: A Geographic Interpretation. New Haven, USA: Yale University Press, 1995.
Dupanloup I, Schneider S, Excoffier L : Inferring the impact of linguistic boundaries on population differentiation and the location of genetic barriers: a new approach; in Renfrew C, Boyle K (eds): Archaeogenetics: DNA and the Population Prehistory of Europe. Cambridge: MacDonald Institute Cambridge, 2000, pp 325–331.
Ruhlen MA : A Guide to the World's Languages, Vol. 1: Classification. Stanford: Stanford University Press, 1991.
Bitima T : A Dictionary of Oromo technical terms Oromo; in Sasse HJ (ed): Cologne, Germany: Rüdiger Köppe publishers, 2000.
Webb VN : African Voices: An Introduction to the Languages and Linguistics of Africa, 1st edn. Cape Town, South Africa: Oxford University press, 2000.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens in extant Europeans: a Y-chromosome perspective. Science 2000; 290: 1155–1159.
Passarino G, Semino O, Magri C et al: The 49a,f haplotype 11 is a new marker of the EU19 lineage that traces migrations from northern regions of the Black Sea. Hum Immunol 62: 922–932. Erratum in Hum Immunol 2001; 62: 1313–1314.
Fattovich R : The Near East and eastern Africa: their interaction; in Vogel JO (ed):: Encyclopaedia of Precolonial Africa: Archaeology, History, Languages, Cultures, and Environments. California: AltaMira Press, Walnut Creek, 1997.
Al-Zahery N, Semino O, Benuzzi G et al: Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 2003; 28: 458–472.
Kayser M, Brauer S, Weiss G et al: Melanesian origin of Polynesian Y chromosomes. Curr Biol 10: 1237–1246. (Erratum in Curr Biol 2000; 11: 141–142).
Kayser M, Brauer S, Weiss G et al: Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet 2003; 72: 281–302.
Acknowledgements
We thank Ms Annemette Holbo Birk for technical assistance and Anders Buchard, Anders Hansen and Bo Simonsen for helpful discussions. The work was supported by grants to JJ Sanchez from Ellen and Aage Andersen's Foundation and by Grant No. PI2000/053 to A Hernandez from Consejería de Educación y Cultura (DGUI), Gobierno de Canarias.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sanchez, J., Hallenberg, C., Børsting, C. et al. High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males. Eur J Hum Genet 13, 856–866 (2005). https://doi.org/10.1038/sj.ejhg.5201390
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201390
Keywords
This article is cited by
-
Genome-wide analyses disclose the distinctive HLA architecture and the pharmacogenetic landscape of the Somali population
Scientific Reports (2020)
-
Prevalence of common hereditary risk factors for thrombophilia in Somalia and identification of a novel Gln544Arg mutation in coagulation factor V
Journal of Thrombosis and Thrombolysis (2017)
-
Genetic differentiation between upland and lowland populations shapes the Y-chromosomal landscape of West Asia
Human Genetics (2017)
-
Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup
Scientific Reports (2016)
-
Mapping human dispersals into the Horn of Africa from Arabian Ice Age refugia using mitogenomes
Scientific Reports (2016)