Abstract
DRONC is an apical Drosophila caspase essential for programmed cell death during fly development. During metamorphosis, dronc gene expression is regulated by the steroid hormone ecdysone, which also regulates the levels of a number of other critical cell death proteins. As DRONC protein levels are important in determining caspase activation and initiation of cell death, we have analyzed the regulation of the dronc promoter using transgenic flies expressing a LacZ reporter gene under the control of the dronc promoter. Our results indicate that dronc expression is highly dynamic during Drosophila development, and is controlled both spatially and temporally. We demonstrate that while a 2.3 kb dronc promoter region contains most of the information required for correct gene expression, a 1.1 kb promoter region is expressed in some tissues and not others. We further demonstrate that during larval–pupal metamorphosis, two ecdysone-induced transcription factors, Broad-Complex and E93, are required for correct dronc expression. Our data suggest that the dronc promoter is regulated in a highly complex manner, and provides an ideal system to explore the temporal and spatial regulation of gene expression driven by nuclear hormone receptors.
Similar content being viewed by others
Introduction
In metazoans, programmed cell death (PCD), often by a morphologically distinct process called apoptosis, is essential for removing unwanted cells.1 The execution phase of PCD requires the activation of a unique class of proteases, termed caspases.2,3 The initial activation of caspases requires specific adaptor molecules that mediate procaspase oligomerization.4,5 Once activated, apical caspases process and activate downstream effector caspases which cleave a large number of cellular proteins in condemned cells to bring about characteristic dismantling of the cell architecture.2,3,6 As key cell death molecules, including caspases, are present in healthy cells, it is generally believed that apoptosis is primarily regulated post-transcriptionally. However, many components of the core apoptosis machinery, including some caspases, are transcriptionally regulated during cell death signaling, suggesting that the levels of the prosurvival and proapoptotic factors in the cell may be crucial to activate the apoptotic program.1 Consistent with this, various signals such as cytotoxic insults, hormones, and growth factors regulate the activation of the death program by controlling the balance between prosurvival and proapoptotic proteins of the core cell death machinery.1 To understand cell death regulation, it is thus essential to understand the role of transcriptional control in the execution of apoptosis.
In Drosophila melanogaster, pulses of the steroid hormone 20-hydroxyecdysone (ecdysone) throughout fly development regulate molting, cell proliferation, differentiation, and PCD in a highly controlled manner.1,7,8 Importantly, during metamorphosis, ecdysone is a primary regulator of PCD in larval tissues such as the salivary glands and midgut.1,9–11 Ecdysone binds to its heterodimeric EcR/Usp receptor, and transcriptionally regulates a number of primary response genes. During the transition of larva into pupa, a late third instar larval (L3) ecdysone pulse signals puparium formation, followed by a second pulse approximately 12 h later which initiates head eversion and pupal development.1,9,10,11 Cell death in the larval midgut begins in response to the late larval ecdysone pulse, whereas salivary glands undergo removal around 15 h later in response to the late prepupal hormone pulse.9 The regulation of PCD by ecdysone is complex and involves EcR/Usp itself and a number of induced transcription factors including βFTZ-F1, BR-C, E74, E75, and E93.1,12,13,14,15,16,17,18,19 Several of these factors regulate the expression of cell death/survival genes, including rpr, hid, dark, dronc, diap1, and diap2.1,12
In Drosophila, the main components of the cell death machinery, including caspases, Bcl-2 homologues, and an Apaf-1 homologue, are conserved.1,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34 Among the seven caspases in Drosophila, DRONC is the main initiator caspase that appears to be functionally similar to CED-3 in Caenorhabditis elegans and caspase-2/caspase-9 in mammals.23,35,36,37 DRONC was the first caspase shown to be regulated by a steroid hormone.23 In the salivary glands and midgut, dronc expression is dramatically upregulated by ecdysone prior to their removal during metamorphosis.23 dronc is also rapidly upregulated by ecdysone in Drosophila l(2)mbn cells, which undergo apoptosis in response to ecdysone treatment.38 Furthermore, DRONC is essential for ecdysone-mediated apoptosis of l(2)mbn cells.38 Although the importance of dronc regulation is well established for ecdysone-mediated cell death, the mechanisms of dronc regulation remain poorly understood. In the present study, we have analyzed the regulation of the dronc promoter, and we report that different regions of the dronc promoter are involved in ecdysone-dependent and -independent temporal and spatial regulation of dronc expression during fly development.
Results
dronc promoter-driven LacZ expression in embryos and adult ovaries
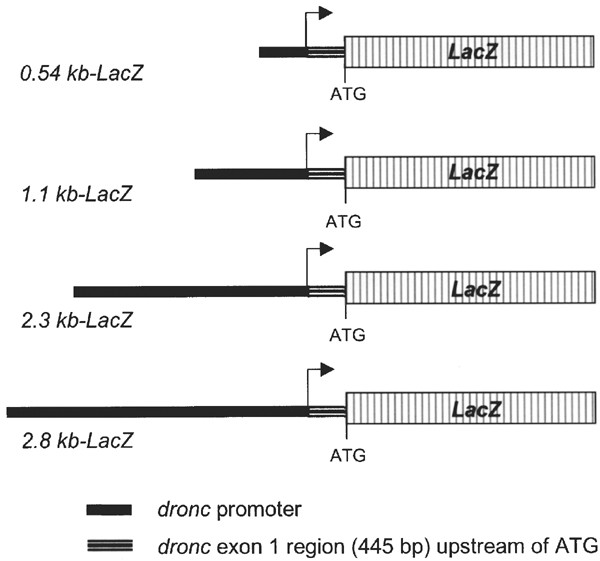
We have previously shown that the dronc transcript is expressed highly in embryos, at very low levels in first instar (L1) to mid-L3 stages, and at higher levels in the midgut and salivary glands from late L3.23 Low levels of dronc expression are also seen throughout the L3 eye discs and in brain lobes, whereas strong expression is evident in adult egg chambers.23 To study the in vivo regulation of dronc expression, we generated a number of transgenic constructs containing various lengths of the dronc promoter region fused to the LacZ gene containing a nuclear localization signal (Figure 1). This allowed us to monitor the expression of the reporter LacZ, the product of which accumulates in the nuclei of cells. Transgenic Drosophila lines containing 0.54, 1.1, 2.3, and 2.8 kb of the dronc promoter region were generated. As the expression patterns of 2.3 and 2.8 kb promoter-LacZ were similar, only the results with 2.8 kb lines, which were analyzed in detail, are presented in this paper. Several lines obtained from the same construct were analyzed to ensure that the reporter expression was not affected due to the transgene integration site.
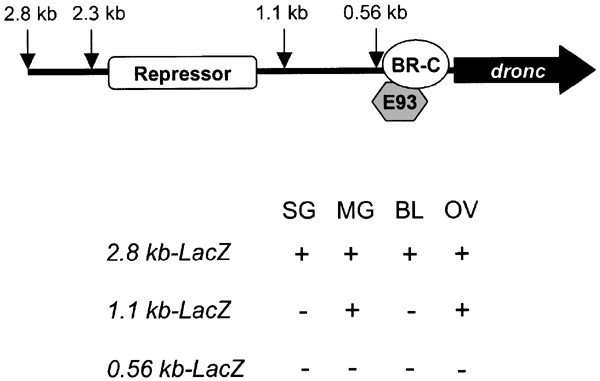
dronc Promoter-LacZ constructs used to generate transgenic flies. Various lengths of the promoter upstream of the dronc transcription start site (shown by an arrow) were cloned upstream of a LacZ reporter gene containing a nuclear localization signal, into the pCaSpeR-4 Drosophila transformation vector. All constructs also contain the 445 bp noncoding region of the dronc exon 1. In all constructs, the initiator ATG codon is the natural dronc initiation codon
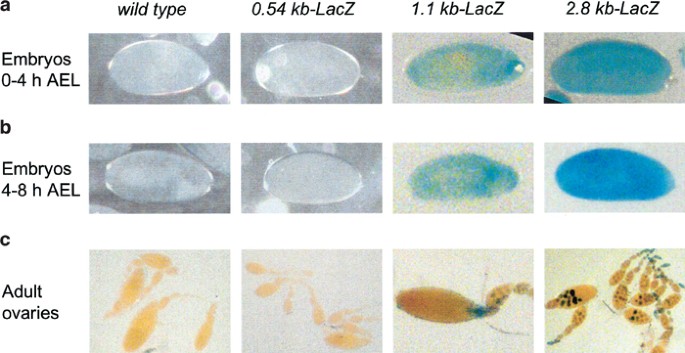
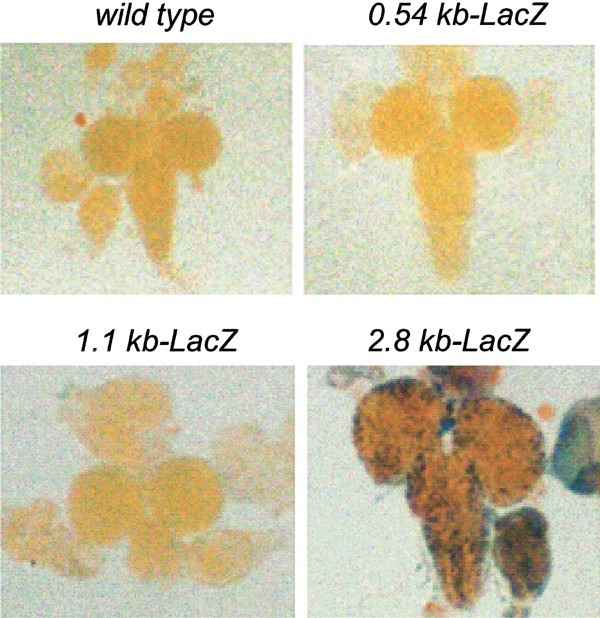
An analysis of β-galactosidase (β-gal) activity in early embryos collected at 0–4 h after egg laying (AEL) stained with X-gal demonstrated that while the 1.1 and 2.8 kb promoter-LacZ lines showed β-gal expression, the 0.54 kb promoter-LacZ lines did not (Figure 2a). The 2.8 kb construct showed a somewhat higher expression of β-gal in embryos 4–8 h AEL (Figure 2a), suggesting that while 1.1 kb may be driving the basal expression of the reporter, 2.8 kb is required for maximal promoter activity upon commencement of the zygotic gene expression. We have previously shown that the dronc transcript is maternally deposited in the early embryos.23 As zygotic expression of genes in Drosophila does not begin until around stage 5 (∼3 h AEL), β-gal expression seen in embryos 0–4 h AEL may largely represent maternally deposited LacZ. Therefore, we tested the activity of β-gal in egg chambers of adult ovaries from transgenic lines. As in embryos, no β-gal expression was evident in lines carrying the 0.54 kb-LacZ construct (Figure 2b). However, both 1.1 and 2.8 kb promoters were able to drive LacZ expression in nurse cells (Figure 2b), suggesting that the 1.1 kb promoter was sufficient for β-gal expression. As nurse cells dump their contents into the developing oocyte, the β-gal activity in 0–4 h embryos with 1.1 and 2.8 kb promoter transgenes may primarily be due to the maternally deposited transcript. As the activity of the 2.8 kb-LacZ transgene is higher in 4–8 h embryos, we conclude that 2.8 kb promoter region is required to direct the limited zygotic expression of dronc in embryos.
Expression of various dronc promoter-LacZ transgenes in wild-type (W1118) or transgenic Drosophila embryos (a, b) and egg chambers (c). In (a, b), the time shown on the left represents h AEL. Dechorionated mixed-stage embryos 0–4 h AEL (a) or 4–8 h AEL (b), or dissected ovaries (c) from adult flies were stained with X-gal, as described in Materials and Methods. After mounting, the stained samples were photographed using an Olympus microscope fitted with a digital camera. β-Gal staining in transgenic flies containing the 1.1 and 2.8 kb promoter transgene appears diffused in embryos (a, b), and as distinct nuclear spots in the nurse cells in the egg chambers (c). In the wild-type and 0.54 kb promoter-LacZ embryos (a, b), no X-gal staining was apparent. Phase-contrast microscopy was used to visualize these unstained embryos. All the reporter lines are homozygous for the transgene. Multiple lines derived from each transgenic construct were analyzed for β-gal activity. The figure shows representative examples of transgene expression
Regulation of dronc promoter in the midgut and salivary glands during metamorphosis
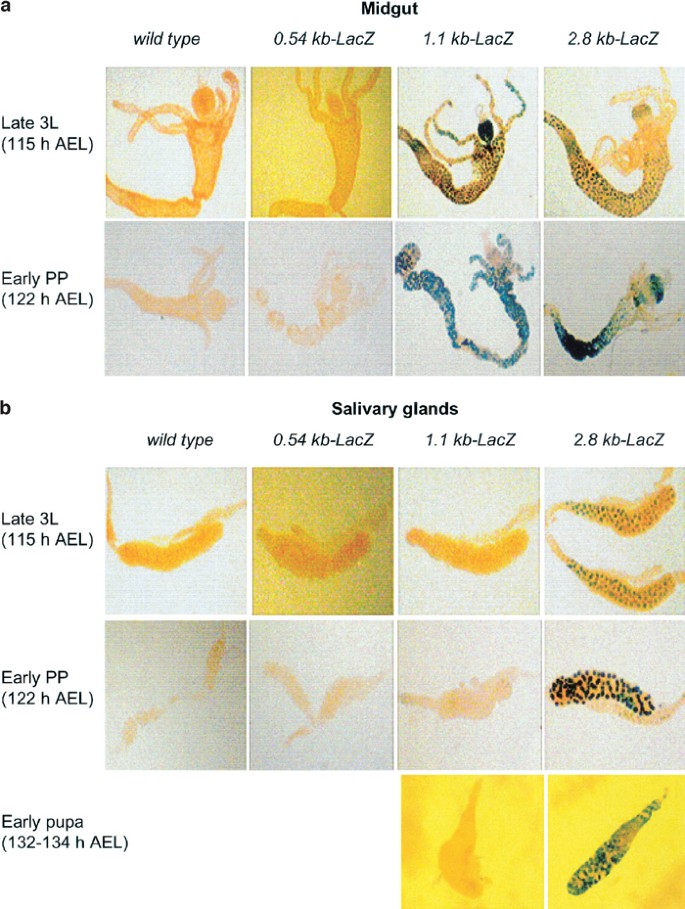
As a major ecdysone-mediated upregulation of dronc expression occurs in the larval salivary glands and midgut just prior to their removal, we analyzed the expression of the promoter-LacZ transgenes in these tissues. In midguts dissected from late L3 (∼115 h AEL) or early prepupae (∼122 h AEL), 1.1 and 2.8 kb promoter-LacZ transgenes were expressed efficiently, however 2.8 kb promoter-driven LacZ expression is lower in gastric caecae (Figure 3a), indicative of different spatial regulation of the two promoter constructs due to heterogeneity in this tissue. The 0.54 kb-LacZ promoter was unable to drive the reporter gene expression (Figure 3a). Note that early prepupal midgut is undergoing PCD at this stage (Figure 3a, lower panel). Interestingly, in salivary glands isolated from late L3 or early prepupae, both 0.54 and 1.1 kb promoter-LacZ transgenes failed to express, however, the 2.8 kb promoter-LacZ was expressed efficiently (Figure 3b, upper two panels). As salivary glands undergo histolysis ∼15 h later than midgut, we also analyzed the expression of the 1.1 and 2.8 kb-LacZ transgenes in salivary glands isolated from early pupae (132–134 h AEL). As in earlier stages, only 2.8 kb promoter was able to drive the reporter expression in salivary glands from this late stage (Figure 3b, lower panel). A 2.3 kb promoter-LacZ transgene showed a pattern of β-gal expression similar to that of the 2.8 kb promoter-LacZ construct (data not shown). These results suggest that while the 1.1 kb promoter is sufficient to drive the expression of dronc in the midgut, at least 2.3 kb of the promoter is required for driving the expression in salivary glands. These results also indicate that a region between 1.1 and 2.3 kb of the promoter is essential for dronc expression in salivary glands. We noted that in salivary glands, reporter expression driven by the 2.8 kb promoter construct was somewhat higher and earlier in development when compared to endogenous dronc expression (data not shown). This observation suggests that additional control elements may be necessary for the precise temporal regulation of dronc expression in salivary glands.
Expression of various dronc promoter-LacZ transgenes in the midgut (a) and salivary glands (b) from the wild-type and transgenic animals. The midgut and salivary glands were dissected out from late L3 (∼115 h AEL) and early prepupae (∼122 h AEL), and stained with X-gal. Salivary glands, which undergo histolysis later than midgut, were also dissected out from an early prepupal stage (132–134 h AEL) and stained with X-gal (bottom panel in (b)). Note that the prepupal midguts are undergoing PCD, whereas salivary glands, which are deleted around 15 h later, are still fully intact at this stage. In the midgut, the prepupal 2.8 kb promoter-LacZ reporter expression pattern is similar to that observed in late L3. No β-gal staining was observed in the midgut and salivary glands from wild-type controls or in 0.54 kb promoter-LacZ transgenic lines
Expression of the promoter-LacZ transgenes in larval brain lobes
Our previous studies have shown that endogenous dronc is expressed in the L3 brain lobes, salivary glands, and midgut.23 Given the spatial pattern of dronc promoter function in the salivary gland and midgut, we further analyzed β-gal expression in the larval brain lobes of promoter-LacZ transgenic lines. As shown in Figure 4, neither 0.54 or 1.1 kb promoter was able to express the reporter in brain lobes, but the 2.8 kb promoter (also the 2.3 kb promoter – data not shown) could efficiently drive LacZ expression. This suggests that, as in larval/prepupal salivary glands, a 2.3 kb promoter region is required for expression in this tissue.
Expression of various dronc promoter-LacZ transgenes in the larval brain lobes. The brain tissue was dissected out from late L3, and stained with X-gal as in Figure 3
Temporal regulation of the dronc promoter
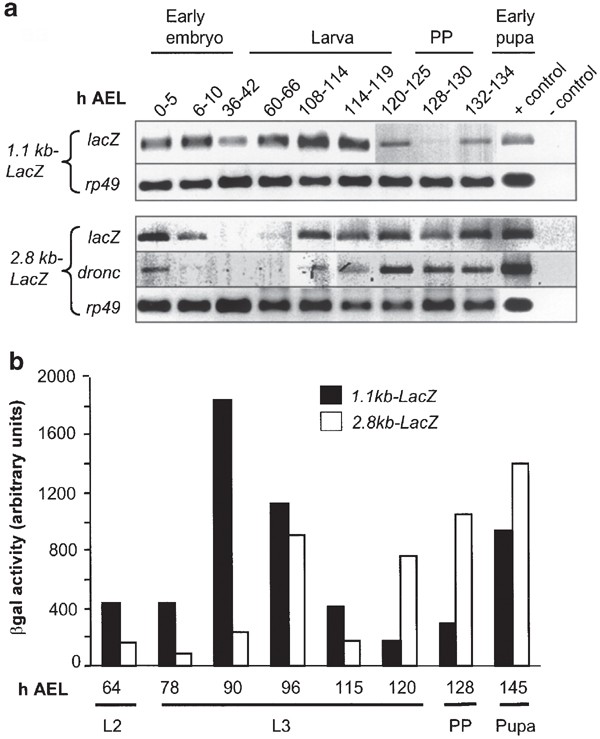
In an effort to ascertain the minimal promoter requirement for correct temporal regulation throughout Drosophila development, we compared expression of the LacZ transgene with that of endogenous dronc. An analysis of dronc and LacZ transcript in whole animals during development suggested that the pattern of the 2.8 kb promoter-driven LacZ expression is similar to that of endogenous dronc (Figure 5a). This suggests that the 2.8 kb of dronc promoter contains most information required for the temporal regulation of dronc expression.
Expression of endogenous dronc and the promoter-LacZ transgenes in animals at various stages of development. In (a), RT-PCR analysis of LacZ, dronc, and rp49 (as a control) expression is shown. The developmental stages are indicated as h AEL. Both the 1.1 and 2.8 kb promoter-LacZ constructs respond to the late L3 and the prepupal ecdysone pulses, which direct endogenous dronc expression. In all, 22–25 cycles of PCR were carried out using cDNA prepared from animals at various stages of development and gene-specific primers. Note that the expression pattern of the 2.8 kb-LacZ transgene is similar to that of endogenous dronc. However, the expression of the 1.1 kb-LacZ transgene is deregulated, and can be clearly seen in the early larval stages when endogenous dronc expression is very low. In prepupal (PP) and early pupal stages, the 1.1 kb-LacZ transgene expression is lower, presumably due to a lack of expression in salivary glands. No expression for the 0.54 kb-LacZ transgene was observed at any stage (data not shown). The positive (plasmid DNA) and negative (−cDNA) PCR controls are shown in the last two lanes of each panel. In (b), β-gal activity was determined in the larvae, PP, and pupae from 1.1 and 2.8 kb-LacZ transgenic animals. Note that the 1.1 kb promoter-driven LacZ expression is much higher in early larval stages. L2, second instar larvae; L3, third instar larvae
We further analyzed the 1.1 and 2.8 kb promoter activity in larval and prepupal stages, by assaying for β-gal activity in whole animals at various stages of development (Figure 5b). Consistent with low dronc expression, the 2.8 kb promoter-LacZ-driven β-gal expression was low in the second instar larvae (L2) which increased significantly at 96 h AEL, followed by a sharp decrease and then steady increase from 120 h AEL, presumably in response to the late larval pulse of ecdysone which punctuates the commencement of metamorphosis. Interestingly, 1.1 kb-LacZ-driven expression of β-gal was somewhat higher in the L2, peaked early at 90 h AEL, and then declined thereafter until the prepupal ecdysone pulse (Figure 5b). The elevated activity of the 1.1 kb promoter at 90 h AEL suggests that this region of the promoter lacks a control mechanism normally required to suppress dronc expression at this stage of development. In other words, our data suggest that the region of dronc promoter between 1.1 and 2.8 kb is regulated by a temporally controlled transcriptional repressor. This is supported by the observation that the 2.8 kb promoter is less responsive to the mid-L3 ecdysone pulse which initiates the larval wandering stage.7
Role of E74A, BR-C, and E93 in the regulation of dronc expression
To analyze the roles of ecdysone-induced genes in regulating dronc expression, we expressed the dronc promoter-LacZ transgenes in a background null for E74A, BR-C, and E93, three EcR/Usp-regulated factors implicated in the regulation of PCD during larval/pupal metamorphosis.11,16,17,18,19 As the midgut is removed about 15 h earlier than salivary glands in response to the late larval ecdysone pulse that initiates pupariation,10,12 it was dissected out from late L3 (∼114 h AEL) and early (white) prepupae (∼120–125 h AEL). Salivary glands, which undergo histolysis in response to the prepupal ecdysone pulse,10,12 were dissected out from late L3, early prepupae, and early pupae (∼132 h AEL).
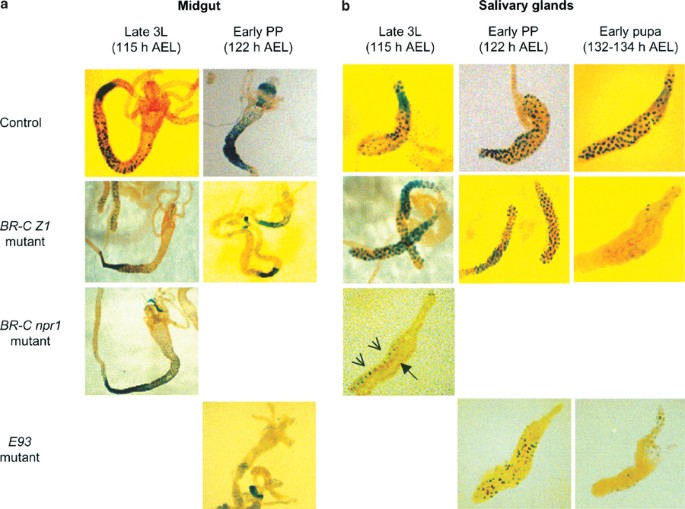
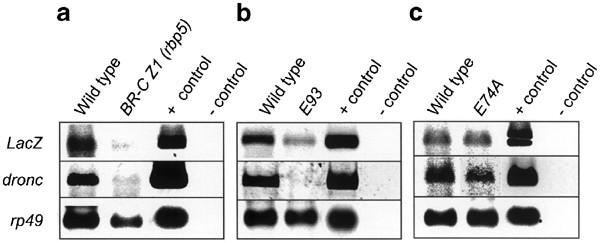
We used two BR-C mutants, npr141 and rbp5.11 npr1 mutants are null for all BR-C isoforms, and fail to initiate pupariation.41 The rbp5 mutant is deficient in the BR-C Z1 isoform, shows a defect in the removal of the larval salivary gland, and fails to develop beyond early pupae.11,39 When the 2.8 kb-LacZ transgene was expressed in the rbp5 mutant background, there was no effect on expression in the midgut at both the late L3 or early prepupal stages (Figure 6a). Similar results were obtained when the 2.8 kb-LacZ transgene was expressed in a BR-C null (npr1) background in the late L3 midguts. Midguts from later stages of development could not be analyzed because npr1 mutants do not develop beyond this stage. Unlike in the midgut, the expression of the 2.8 kb-LacZ transgene was severely reduced in salivary glands from early pupal stages in the rbp5 mutant and late larval stages in the npr1 mutant (Figures 6b, 7a). These data suggest that BR-C is essential for proper expression of dronc in salivary glands, but not in the midgut.
Expression of various dronc promoter-LacZ transgenes in the midgut (a) and salivary glands (b), from BR-C and E93 mutants. The 2.8 kb promoter-LacZ transgene was expressed in a wild-type (control), BR-C (rbp5 or npr1) or E93 null background, as described in Materials and Methods. The midgut and salivary glands were dissected out from late 3L (∼115 h AEL) and early prepupae (PP) (∼122 h AEL), and stained with X-gal. Salivary glands were also dissected out from the early pupal stage (132–134 h AEL) animals, and stained with X-gal. β-Gal staining seen in the BR-C npr1 salivary gland (b, late 3L) represents fat body (arrow heads), while the salivary gland (arrow) itself is not stained. In E93 mutants, staining in both the midgut and salivary glands is reduced, and can only be seen in long exposures (bottom panels in (a) and (b))
RT-PCR analysis of LacZ and dronc expression in the salivary glands of 2.8 kb promoter-LacZ transgenic flies in BR-C Z1 (a), E93 (b), and E74A (c) null background. Salivary glands were dissected out from animals around 130–133 h AEL. Rp49 expression was used as a control. The promoter-LacZ transgene (2.8 kb) was in either the wild-type (W1118) or mutant background as indicated. The positive (plasmid DNA) and negative (−cDNA) PCR controls are shown in the last two lanes of each panel
The E93 gene encodes a novel nuclear protein, and is specifically required for ecdysone-induced cell death in both the midgut and salivary glands.19 In E93 null mutants, larval midgut and salivary glands persist into early pupal stages when development is arrested.19 When the 2.8 kb-LacZ transgene was introduced into the E93 mutant background, the expression of β-gal was strongly suppressed in all regions of the midgut and salivary glands at the time prior to the histolysis of these tissues (Figures 6, 7b). These data indicate that E93 is required for the maximal expression of dronc during PCD in both tissues.
We also analyzed the effect of E74A mutants on 2.8 kb-LacZ transgene expression. Consistent with previous findings,39 we observed no significant qualitative or quantitative changes in X-gal-stained salivary glands (data not shown). Likewise, RT-PCR showed 2.8 kb-LacZ transgene activity in E74A-deficient salivary glands comparable to controls (Figure 7c, top panel). Although the endogenous dronc transcript appears to be somewhat reduced in the E74A-deficient salivary glands in Figure 7c (middle panel), this was not always seen. These data suggest that E74A does not play a crucial role in regulating dronc expression during salivary gland cell death.
Discussion
DRONC is the only CARD containing apical caspase in Drosophila and is predicted to be the main initiator caspase functionally similar to CED-3 in C. elegans and caspase-2/caspase-9 in mammals.23,37 RNAi studies suggest that DRONC is essential for PCD during embryogenesis37 and for ecdysone-induced cell death.38 As evident from this study and previous observations,23 the expression of dronc transcript during development is highly dynamic. As intracellular levels of DRONC protein may determine the sensitivity of cells to undergo PCD, it is important to understand how dronc expression is regulated during development. As summarized in Figure 8, the results reported in this paper provide the first analysis of the dronc promoter, and form a basis for further studies. Given that dronc is regulated both temporally and spatially and dronc gene expression, at least during metamorphosis, is regulated by the steroid hormone ecdysone, the dronc promoter provides a convenient model system for dissecting out the components of the regulatory apparatus mediating gene expression by nuclear hormones.
Summary of the dronc promoter analysis. Potential BR-C and E93-binding elements and repressor-binding site are indicated. The lower panel summarizes the results of dronc promoter-LacZ expression in various tissues. SG, larval or prepupal salivary glands; MG, larval or prepupal midgut; BL, larval brain lobes; OV, adult ovaries
Experiments outlined in this paper demonstrate that 2.3 kb of the dronc promoter is largely sufficient for temporal expression (compared to endogenous dronc) throughout development. Previous experiments have shown that dronc is predominantly expressed in the larval and prepupal salivary glands and midgut, and larval brain lobes.23 We have shown that 2.3 kb of the dronc promoter contains all necessary elements for correct spatial regulation of dronc expression in these tissues.
In order to identify transcription factors responsible for both temporal and spatial regulation of dronc and ecdysone-mediated PCD, it is of vital importance to elucidate the regions of the promoter essential for dronc expression in different tissues. In addition, it would be of interest to determine if there is a single promoter region controlling the spatial expression profile of dronc, or if different promoter regions are required in different tissues. LacZ transgenic reporter experiments reveal that the 2.3 kb promoter is the minimal requirement for correct expression in brain lobes and salivary glands. Furthermore, the region between 1.1 and 2.3 kb contains transcription factor-binding sites essential for expression in these tissues. This region also seems to harbor a repressor element important to keep dronc levels low during periods when ecdysone titers are low. Surprisingly, regulation of dronc transcription is markedly different in the midgut. The region between 1.1 and 2.3 kb is not important for transcription in this tissue, as 1.1 kb of the promoter is sufficient for expression. These results clearly demonstrate that distinct regions of the promoter are required for expression in different tissues, and implies that different transcription factors regulate dronc expression in a tissue-dependent manner.
The two ecdysone-induced transcription factors BR-C and E93 are essential for dronc expression in salivary glands.1,12,19,39 In the midgut, however, only E93 seems to be important.40 Our results of dronc promoter-LacZ transgenic expression in flies deficient in BR-C and E93 are consistent with recent findings.19,38,39,40 LacZ expression driven by the 2.8 kb promoter is severely impaired in salivary glands of BR-C (rbp5 and npr) or E93 mutants, whereas expression is impaired only in the midgut of E93 mutant background animals. This further supports the idea that the mechanisms governing dronc regulation are tissue specific. The key questions arising from these experiments are: why does the BR-C Z1 isoform (rbp5 mutant) regulate dronc in the salivary glands and not in the midgut? What factors are binding to the 1.1–2.3 kb region of the promoter in salivary glands, and why are they not as important in the midgut? Our previous results show that either BR-C Z1- or BR-C Z1-regulated proteins bind to the dronc proximal promoter (7–176 bp) and control its expression.38 Transactivation of the 2.8 kb promoter by BR-C Z1, however, was only seen in specific cell types.38 Given that BR-C Z1 is also expressed in the midgut, this implies that it may be acting through cofactors which are not expressed in the midgut, yet are specifically recruited to the dronc promoter. Alternatively, BR-C Z1 induces the expression of another factor which binds to the promoter, and this factor is absent in the midgut.
Since we have shown that the proximal promoter alone (0.54 kb) is not sufficient for expression in the salivary gland, we believe that BR-C Z1 (or a Z1-regulated protein) is cooperating with other transcription factors binding upstream (1.1–2.3 kb), which are essential for salivary gland expression. We have found that E93 acts through the first 600 bp of the dronc promoter by transactivation studies (DC and SK, unpublished data); however, no direct binding of E93 to the dronc (or any other) promoter has been shown so far. Additionally, a preliminary analysis indicates the presence of an EcR/Usp-binding site between 1.1 and 2.3 kb of the dronc promoter, and in vitro experiments show that this element may be important in regulating dronc expression (DC, TD, and SK, unpublished data). Since the proximal promoter (0.54 kb) alone is not sufficient for expression, cooperation of BR-C and E93 with EcR/Usp and other unknown factors may be important for temporal and spatial regulation of dronc expression during development. Identification of these factors will be important for fully understanding dronc transcription during development.
Overall, we have established the minimal dronc promoter requirement for spatial and temporal expression to be within the 2.3 kb region upstream of the dronc gene. This region is important for both BR-C- and E93-mediated transcription in salivary glands and E93 transcription in the midgut. Importantly, the 1.1–2.3 kb promoter region harbors elements important for salivary gland expression and a putative repressor element. The 0.54–1.1 kb promoter region is important for expression in the midgut. These regions will form the basis of future experiments designed to identify factors necessary for the regulation of dronc expression during PCD.
Materials and Methods
Transgenic and mutant stocks and crosses
All dronc promoter-nuclear LacZ reporter constructs were generated by double-insert ligations into the pCasper-4 p-element-mediated transformation vector,41 in the following manner. Variable promoter lengths were PCR amplified from D. melanogaster genomic DNA using a common reverse primer containing an Nco1 site at the ATG of dronc. 5′ blunt and 3′ Nco1 dronc promoter fragments were ligated with 5′ Nco1 and 3′ Not1-cut LacZ into pCasper-4 with a Klenow end-filled EcoR1 and a Not1 site.
Transgenic flies were generated and maintained as previously described.42 Briefly, dronc promoter-LacZ reporter germline transformation constructs (∼500 ng/μl) and transposase helper plasmid Δ2–3 (∼150 ng/μl) were microinjected into precellularized Drosophila W1118 embryos. w+ transformants were screened by eye color, mapped, and homozygous stocks generated by established techniques.
For BR-C Z1 isoform mutant (rbp5)11 experiments, males homozygous for the second or third chromosome dronc promoter-LacZ transgenes were crossed to virgin yrbp5 heterozygote females balanced with the sex chromosome balancer Binsn. The yellow mouthhook male larvae, prepupae, and early prepupae (hemizygous for the mutated BR-C gene and heterozygous for the reporter) were selected for analysis. Control flies are male larvae with normal mouthhooks, which have wild-type BR-C genetic backgrounds (Binsn/Y). A similar crossing strategy was employed to analyze reporter activity in the BR-C null mutant npr1.43
Fly stocks homozygous for a second chromosome 2.8 kb dronc promoter-LacZ transgene and heterozygous for TM6B balancer chromosome, and either the E93 mutant19 or deficiency (Df[3R]93Fx2), were crossed, and non-TM6B progeny (DrPr2.8kb-LacZ; E93′/Df[3R]93Fx2) compared to control TM6B progeny (DrPr2.8kb-LacZ; E931 or Df[3R]93Fx2/TM6B). A similar crossing strategy was undertaken for DrPr2.8kb-LacZ, E74A mutant analysis, employing the mutant E74APneo 16 and E74A deficiency (Df[3L]st-8IK19).
Staging of animals
Developmental stages from the embryos to L3 stages were acquired by ageing animals from the time of egg deposition on grape agar plates to the desired stages at 25°C. L3 stages were determined by the gut-clearance technique following the growth of animals on bromophenol blue supplemented food.44 Prepupal and pupal stages were attained by collecting newly pupariated animals from clear-gutted L3 populations every 30 min, and ageing at 25°C to desired stages before collection and analysis.
For mutant and dronc promoter–reporter crosses, approximately 50 virgins and males were left for 3 days in vials containing standard cornmeal media, and then transferred to grape agar lay tubes for staging lays. Animals were collected, rinsed briefly in PBS, frozen in liquid nitrogen, and stored at −70°C for assaying or RNA extraction, or dissected and stained as below.
Detection of β-gal expression
β-Gal detection in embryos was carried out essentially as described.45 Briefly, the embryos were collected from grape agar lay plates, dechorionated in 50% bleach/50% PBT, washed in water and fixed for 20 min in 50% heptane/50% fixative (0.1 M sodium phosphate, pH 7.5/4% formaldehyde) while nutating. Embryos were rehydrated with 0.7% NaCl/0.04% Triton X-100 for 5 min, stained with X-gal solution (5 mM K4[Fe(CN)6]/5 mM K3[Fe(CN)6]/0.2% X-gal stock (20 mg/ml) in PBS), and incubated at 37°C for a period dependent on transgene expression levels (usually overnight). Embryos were devitellinized with 50% heptane/50% methanol by vortexing for 20 s, washed with ethanol and mounted in 80% glycerol, 20% PBT for analysis. Adult, pupal, and larval tissues were stained as described.45 Briefly, the tissues were dissected in PBS, fixed for 10 min in 2.5% gluteraldehyde/50 mM Pipes (pH 7.5), washed 3 × 5 min in PBS, and incubated at 37°C in X-gal staining solution. Tissues were washed in PBS, incubated overnight in 80% glycerol/20% PBS, mounted and photographed.
Quantitative β-gal assays
Quantitation of β-gal activity was carried out essentially as described,45 with some modifications. In all, 5–10 animals were homogenized in 100 μl assay buffer (50 mM potassium phosphate/1 mM MgCl2, pH 7.5), the volume made up to 1 ml and vortexed briefly. A volume of 10 μl was transferred to 200 μl of chlorophenol red β-D-galactopyranoside (CPRG) solution (1 mM CPRG final concentration), and incubated at 37°C. β-Gal activity is represented as the change in A574 over 2 h, divided by the sample protein concentration, and then multiplied by an arbitrary constant. Control values from W1118 samples were subtracted to account for endogenous β-gal activity.
RT-PCR
Total RNA was extracted from the dissected tissues and whole animals using Trizol reagent (Invitrogen), according to the manufacturer's protocol. Up to 5 μg of total RNA was used as template for cDNA synthesis, in a 15 μl reaction with 200 ng of oligo d(T)18 primer, using a First-Strand Synthesis Kit (Amersham), according to the manufacturer's protocol. Using 2 μl of cDNA template, PCR amplification was performed using appropriate primers in a 50 μl reaction employing 22–25 cycles. Drosophila Rp49 was used as a control. PCR reaction (10 μl) was electrophoresed on a 1.5% agarose gel for analysis.
Abbreviations
- PCD:
-
programmed cell death
- β-gal:
-
β-galactosidase
- CPRG:
-
chlorophenol red β-d-galactopyranoside
- X-gal:
-
5-bromo-4-chloro-3-indolyl-β-d-galactoside
- AEL:
-
after egg laying
- L2:
-
second instar larvae
- L3:
-
third instar larvae
- PP:
-
prepupae
References
Baehrecke EH (2002) How death shapes life during development. Nat. Rev. Mol. Cell. Biol. 3: 779–787
Nicholson DW (1999) Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6: 1028–1042
Budihardjo I, Oliver H, Lutter M, Luo X and Wang X (1999) Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15: 269–290
Kumar S (1999) Mechanisms mediating caspase activation in cell death. Cell Death Differ. 6: 1060–1066
Shi Y (2002) Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9: 459–470
Fischer U, Janicke RU and Schulze-Osthoff K (2003) Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10: 76–100
Riddiford LM (1993) In The Development of Drosophila melanogaster. Vol. II, Bate M and Martinez-Arias A (eds) (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 899–939
Truman JW and Riddiford LM (2002) Endocrine insights into the evolution of metamorphosis in insects. Annu. Rev. Entomol. 47: 467–500
Thummel CS (1996) Files on steroids – Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 12: 306–310
Jiang C, Baehrecke EH and Thummel CS (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683
Restifo LL and White K (1992) Mutations in a steroid hormone-regulated gene disrupt the metamorphosis of internal tissues in Drosophila: salivary glands, muscle, and gut. Roux's Arch. Dev. Biol. 201: 221–234
Baehrecke EH (2000) Steroid regulation of programmed cell death during Drosophila development. Cell Death Differ. 7: 1057–1062
Broadus J, McCabe JR, Endrizzi B, Thummel CS and Woodard CT (1999) The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell 3: 143–149
Karim FD, Guild GM and Thummel CS (1993) The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118: 977–988
DiBello PR, Withers DA, Bayer CA, Fristrom JW and Guild GM (1991) The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129: 385–397
Fletcher JC, Burtis KC, Hogness DS and Thummel CS (1995) The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development 121: 1455–1465
Jiang C, Lamblin A-FJ, Steller H and Thummel CS (2000) A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5: 445–455
Lee C-Y and Baehrecke EH (2001) Steroid regulation of autophagic programmed cell death during development. Development 128: 1443–1455
Lee C-Y, Wendel DP, Reid P, Lam G, Thummel CS and Baehrecke EH (2000) E93 directs steroid-triggered programmed cell death in Drosophila. Mol. Cell 6: 433–443
Song Z, McCall K and Steller H (1997) DCP-1, a Drosophila cell death protease essential for development. Science 275: 536–540
Chen P, Rodriguez A, Erskine R, Thach T and Abrams JM (1998) Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev. Biol. 201: 202–216
Fraser AG and Evan GI (1997) Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO J. 16: 2805–2813
Dorstyn L, Colussi PA, Quinn LM, Richardson H and Kumar S (1999) DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA 96: 4307–4312
Dorstyn L, Read SH, Quinn LM, Richardson H and Kumar S (1999) DECAY, a novel Drosophila caspase related to mammalian caspase-3 and caspase-7. J. Biol. Chem. 274: 30778–30783
Doumanis J, Quinn L, Richardson H and Kumar S (2001) STRICA, a novel Drosophila melanogaster caspase with an unusual serine/threoninerich prodomain, interacts with DIAP1 and DIAP2. Cell Death Differ. 8: 387–394
Harvey N, Daish T, Quinn L, Read S, Richardson H and Kumar S (2001) Characterization of the Drosophila caspase, DAMM. J. Biol. Chem. 276: 25342–25350
Colussi PA, Quinn LM, Huang DCS, Coombe M, Read SH, Richardson H and Kumar S (2000) Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J. Cell Biol. 148: 703–710
Igaki T, Kanuka H, Inohara N, Sawamoto K, Nunez G, Okano H and Miura M (2000) Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc. Natl. Acad. Sci. USA 97: 662–667
Baker Brachmann C, Jassim OW, Wachsmuth BD and Cagan RL (2000) The Drosophila bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr. Biol. 10: 547–550
Quinn L, Coomb M, Mills K, Daish T, Coluss P, Kumar S and Richardson H (2003) Buffy, a Drosophila Bcl-2 related protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22: 3568–3579
Rodriguez A, Oliver H, Zou H, Chen P, Wang X and Abrams JM (1999) Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1: 272–279
Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H and Miura M (1999) Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol. Cell 4: 757–769
Zhou L, Song Z, Tittel J and Steller H (1999) HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell 4: 745–755
Richardson H and Kumar S (2002) Death to flies: Drosophila as a model system to study programmed cell death. J. Immun. Methods 265: 21–38
Kumar S and Doumanis J (2000) The fly caspases. Cell Death Differ. 7: 1039–1044
Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA and Kumar S (2002) The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 156: 1089–1098
Quinn LM, Dorstyn L, Mills K, Colussi PA, Chen P, Coombe M, Abrams J, Kumar S and Richardson H (2000) An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J. Biol. Chem. 275: 40416–40424
Cakouros D, Daish T, Martin D, Baehrecke EH and Kumar S (2002) Ecdysone-induced expression of the caspase DRONC during hormone-dependent programmed cell death in Drosophila is regulated by Broad-Complex. J. Cell Biol. 157: 985–995
Lee C-Y, Simon CR, Woodard CT and Baehrecke EH (2002) Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252: 138–148
Lee C-Y, Cooksey BA and Baehrecke EH (2002) Steroid regulation of midgut cell death during Drosophila development. Dev. Biol. 250: 101–111
Thummel CS, Bulet AM and Lipshitz HD (1988) Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 7: 445–456
Brand AH and Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415
Belyaeva ES, Aizenzon MG, Semeshin VF, Kiss II, Koczka K, Baritcheva EM, Gorelova TD and Zhimulev IF (1980) Cytogenetic analysis of the 2B3-4-2B11 region of the X-chromosome of Drosophila melanogaster. I. Cytology of the region and mutant complementation groups. Chromosoma 81: 281–306
Maroni G and Stamey SC (1983) Use of blue food to select synchronous, late third instar larvae. Drosophila Inf. Serv. 59: 142–143
Hazelrigg T (2000) In Drosophila Protocols Sullivan W, Ashburner M and Hawley RS (eds) (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 313–343
Acknowledgements
This work was supported by the National Health and Medical Research Council of Australia. TJD was supported by a Dawes Postgraduate Award from the Royal Adelaide Hospital. We are grateful to Eric Baehrecke, Carl Thummel, and Drosophila Stock Center for fly stocks.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by E Baehreckle
Rights and permissions
About this article
Cite this article
Daish, T., Cakouros, D. & Kumar, S. Distinct promoter regions regulate spatial and temporal expression of the Drosophila caspase dronc. Cell Death Differ 10, 1348–1356 (2003). https://doi.org/10.1038/sj.cdd.4401312
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401312