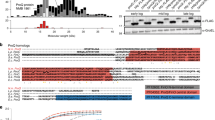
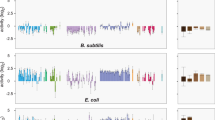
Abstract
Two recent reports have indicated that the H-NS protein in Salmonella enterica serovar Typhimurium has a key role in selectively silencing the transcription of large numbers of horizontally acquired AT-rich genes, including those that make up its major pathogenicity islands. Broadly similar conclusions have emerged from a study of H-NS binding to DNA in Escherichia coli. How do these findings affect our view of H-NS and its ability to influence bacterial evolution?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Dorman, C. J. H-NS: a universal regulator for a dynamic genome. Nature Rev. Microbiol. 2, 391–400 (2004).
Lucchini, S. et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81 (2006).
Navarre, W. W. et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella enterica Sv. Typhimurium. Science 313, 236–238 (2006).
Oshima, T., Ishikawa, S., Kurokawa, K., Aiba, H. & Ogasawara, N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13, 141–153 (2006).
Rimsky, S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7, 109–114 (2004).
Tendeng, C. & Bertin, P. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 11, 511–518 (2003).
Dame, R. T., Wyman, C. & Goosen, N. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83, 231–234 (2001).
Dame R. T., Wyman, C., Wurm, R., Wagner, R. & Goosen, N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277, 2146–2150 (2002).
Kozobay-Avraham, L., Hosid, S. & Bolshoy, A. Curvature distribution in prokaryotic genomes. In Silico Biol. 4, 361–375 (2004).
Olivares-Zavaleta, N., Jauregui, R. & Merino, E. Genome analysis of Escherichia coli promoter sequences evidences that DNA static curvature plays a more important role in gene transcription than has previously been anticipated. Genomics 87, 329–337 (2006).
Pedersen, A. G., Jensen, L. J., Brunak, S., Staerfeldt, H. H. & Ussery, D. W. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 299, 907–930 (2000).
Rimsky, S., Zuber, F., Buckle, M. & Buc, H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42, 1311–1323 (2001).
Groisman, E. A. & Ochman, H. How Salmonella became a pathogen. Trends Microbiol. 5, 343–349 (1997).
Ochman, H., Lawrence, J. G. & Groisman, E. A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000).
Buchrieser, C. et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38, 760–771 (2000).
Dorman, C. J. 2004. Virulence gene regulation in Shigella. In Escherichia coli and Salmonella: Cellular and Molecular Biology. (eds Curtiss, R. et al.) EcoSal[online]. (American Society for Microbiology, Washington DC, 2004).
Brussow, H., Canchaya, C. & Hardt, W. D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602 (2004).
Nye, M. B., Pfau, J. D., Skorupski, K. & Taylor, R. K. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182, 4295–4303 (2000).
Park, K. S., Arita, M., Iida, T. & Honda, T. vpaH, a gene encoding a novel histone-like nucleoid structure-like protein that was possibly horizontally acquired, regulates the biogenesis of lateral flagella in trh-positive Vibrio parahaemolyticus TH3996. Infect. Immun. 73, 5754–5761 (2005).
Dole, S., Nagarajavel, V. & Schnetz, K. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52, 589–600 (2004).
Gowrishankar, J. & Manna, D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica 97, 363–378 (1996).
Rajkumari, K., Kusano, S., Ishihama, A., Mizuno, T. & Gowrishankar, J. Effects of H-NS and potassium glutamate on σS- and σ70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J. Bacteriol. 178, 4176–4181 (1996).
Chen, C. C. & Wu, H. Y. LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J. Biol. Chem. 280, 15111–15121 (2005).
Pflum, M. K. H-NS gives invading DNA the silent treatment. Nature Chem. Biol. 2, 400–401 (2006).
Beloin, C. & Dorman, C. J. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47, 825–838 (2003).
Le Gall, T. et al. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151, 951–962 (2005).
Olekhnovich, I. N. & Kadner, R. J. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357, 373–386 (2006).
Babu, M. M., Teichmann, S. A. & Aravind, L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 358, 614–633 (2006).
Akbar, S., Schechter, L. M., Lostroh, C. P. & Lee, C. A. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47, 715–728 (2003).
Bajaj, V., Lucas, R. L., Hwang, C. & Lee, C. A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22, 703–714 (1996).
Baxter, M. A., Fahlen, T. F., Wilson, R. L. & Jones, B. D. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71, 1295–1305 (2003).
Cirillo, D. M., Valdivia, R. H., Monack, D. M. & Falkow, S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30, 175–188 (1998).
Garmendia, J., Beuzon, C. R., Ruiz-Albert, J. & Holden, D. W. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149, 2385–2396 (2003).
Hensel, M. et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30, 163–174 (1998).
Olekhnovich, I. N. & Kadner, R. J. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 4148–4160 (2001).
Schechter, L. M. & Lee, C. A. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40, 1289–1299 (2001).
Grob, P. & Guiney, D. G. In vitro binding of the Salmonella dublin virulence plasmid regulatory protein SpvR to the promoter regions of spvA and spvR. J. Bacteriol. 178, 1813–1820 (1996).
Robbe-Saule, V., Schaeffer, F., Kowarz, L. & Norel, F. Relationships between H-NS, σS, SpvR and growth phase in the control of spvR, the regulatory gene of the Salmonella plasmid virulence operon. Mol. Gen. Genet. 256, 333–347 (1997).
Sheehan, B. J. & Dorman, C. J. In vivo analysis of the interactions of the LysR-like regulator SpvR with the operator sequences of the spvA and spvR virulence genes of Salmonella typhimurium. Mol. Microbiol. 30, 91–105 (1998).
O'Byrne, C. P. & Dorman, C. J. Transcription of the Salmonella typhimurium spv virulence locus is regulated negatively by the nucleoid-associated protein H-NS. FEMS Microbiol. Lett. 121, 99–105 (1994).
Schechter, L. M., Jain, S., Akbar, S. & Lee, C. A. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71, 5432–5435 (2003).
Dorman, C. J. & Deighan, P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13, 179–184 (2003).
Hillebrand, A., Wurm, R., Menzel, A. & Wagner, R. The seven E. coli ribosomal RNA operon upstream regulatory regions differ in structure and transcription factor binding efficiencies. Biol. Chem. 386, 523–534 (2005).
Prosseda, G. et al. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51, 523–537 (2004).
Schneider, D. A., Ross, W. & Gourse, R. L. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6, 151–156 (2003).
Kelly, A. et al. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150, 2037–2053 (2004).
Grainger, D. C., Hurd, D., Goldberg, M. D. & Busby, S. J. W. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34, 4642–4652 (2006).
Shin, M. et al. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 19, 2388–2398 (2005).
Free, A. & Dorman, C. J. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol. Microbiol. 18, 101–113 (1995).
Hinton, J. C. D. et al. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6, 2327–2337 (1992).
Schröder, O. & Wagner, R. The bacterial regulatory protein H-NS — a versatile modulator of nucleic acid structures. Biol. Chem. 383, 945–960 (2002).
Amit, R., Oppenheim, A. B. & Stavans, J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 84, 2467–2473 (2003).
Augustyn, K. E., Wojtuszewski, K., Hawkins, M. E., Knutson, J. R. & Mukerji, I. Examination of the premelting transition of DNA A-tracts using a fluorescent adenosine analogue. Biochemistry 45, 5039–5047 (2006).
Ussery, D. W., Higgins, C. F. & Bolshoy, A. Environmental influences on DNA curvature. J. Biomol. Struct. Dyn. 16, 811–823 (1999).
Lucas, R. L. & Lee, C. A. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183, 2733–2745 (2001).
Rhen, M. & Dorman, C. J. Hierarchical gene regulators adapt Salmonella enterica to its host milieus. Int. J. Med. Microbiol. 295, 487–502 (2005).
Dorman, C. J., Hinton, J. C. D. & Free, A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7, 124–128 (1999).
Free, A., Porter, M. E., Deighan, P. & Dorman, C. J. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42, 903–917 (2001).
Johansson, J., Eriksson, S., Sondén, B., Wai, S. N. & Uhlin, B. E. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 183, 2343–2347 (2001).
Williams, R. M., Rimsky, S. & Buc, H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178, 4335–4343 (1996).
Zhang, A., Rimsky, S., Reaban, M. E., Buc, H. & Belfort, M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15, 1340–1349 (1996).
Muller, C. M. et al. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188, 5428–5438 (2006).
Esposito, D. et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324, 841–850 (2002).
Bloch, V. et al. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nature Struct. Biol. 10, 212–218 (2003).
Ono, S. et al. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391, 203–213 (2005).
Dame, R. T., Noom, M. C. & Wuite, G. J. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444, 387–390 (2006).
Cheung, K. J., Badarinarayana, V., Selinger, D. W., Janse, D. & Church, G. M. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 13, 206–215 (2003).
Lacour, S. & Landini, P. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195 (2004).
Zhou, Y. & Gottesman, S. Modes of regulation of RpoS by H-NS. J. Bacteriol. 188, 7022–7025 (2006).
Dame, R. T. et al. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 187, 1845–1848 (2005).
Pingoud, A., Fuxreiter, M., Pingoud, V. & Wende, W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62, 685–707 (2005).
Doyle, M. et al. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science in the press.
Cerdan, R. et al. Crystal structure of the N-terminal dimerization domain of VicH, the H-NS-like protein of Vibrio cholerae. J. Mol. Biol. 334, 179–185 (2003).
Atlung T. & Ingmer, H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24, 7–17 (1997).
Nye, M. B. & Taylor, R. K. Vibrio cholerae H-NS domain structure and function with respect to transcriptional repression of ToxR regulon genes reveals differences among H-NS family members. Mol. Microbiol. 50, 427–444 (2003).
Stella, S., Falconi, M., Lammi, M., Gualerzi, C. O. & Pon, C. L. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 355, 169–174 (2006).
Stella, S., Spurio, R., Falconi, M., Pon, C. L. & Gualerzi, C. O. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 24, 2896–2905 (2005).
Ueguchi, C., Seto, C., Suzuki, T. & Mizuno, T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 274, 145–151 (1997).
Owen-Hughes, T. A. et al. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71, 255–265 (1992).
Dersch, P., Schmidt, K. & Bremer, E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8, 875–889 (1993).
Ueguchi, C., Kakeda, M. & Mizuno, T. Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein: H-NS functions as a repressor of its own transcription. Mol. Gen. Genet. 236, 171–178 (1993).
Sondén, B. & Uhlin, B. E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15, 4970–4980 (1996).
Free, A. & Dorman, C. J. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179, 909–918 (1997).
Deighan, P., Beloin, C. & Dorman, C. J. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol. Microbiol. 48, 1401–1416 (2003).
Falconi, M., Higgins, N. P., Spurio, R., Pon, C. L. & Gualerzi, C. O. Expression of the gene encoding the major bacterial nucleotide protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 10, 273–282 (1993).
La Teana, A. et al. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl Acad. Sci. USA 88, 10907–10911 (1991).
Lease, R. A. & Belfort, M. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38, 667–672 (2000).
Acknowledgements
I thank Niamh Ní Bhriain for insightful comments on the manuscript and I am grateful to Jay Hinton, Ferric Fang, William Navarre and Marie Doyle for helpful discussions. Work in the author's laboratory is supported by grants from the Science Foundation Ireland and the Wellcome Trust.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Rights and permissions
About this article
Cite this article
Dorman, C. H-NS, the genome sentinel. Nat Rev Microbiol 5, 157–161 (2007). https://doi.org/10.1038/nrmicro1598
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1598
This article is cited by
-
Transcription-driven DNA supercoiling counteracts H-NS-mediated gene silencing in bacterial chromatin
Nature Communications (2024)
-
c-di-GMP inhibits the DNA binding activity of H-NS in Salmonella
Nature Communications (2023)
-
Xenogeneic silencing strategies in bacteria are dictated by RNA polymerase promiscuity
Nature Communications (2022)
-
Pat- and Pta-mediated protein acetylation is required for horizontally-acquired virulence gene expression in Salmonella Typhimurium
Journal of Microbiology (2022)
-
Regulation of gene expression by protein lysine acetylation in Salmonella
Journal of Microbiology (2020)