Abstract
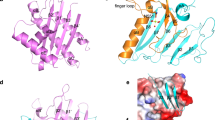
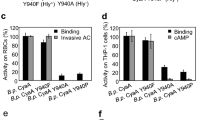
MARTX toxins modulate the virulence of a number of Gram-negative Vibrio species. This family of toxins is defined by the presence of a cysteine protease domain (CPD), which proteolytically activates the Vibrio cholerae MARTX toxin. Although recent structural studies of the CPD have uncovered a new allosteric activation mechanism, the mechanism of CPD substrate recognition or toxin processing is unknown. Here we show that interdomain cleavage of MARTXVc enhances effector domain function. We also identify the first small-molecule inhibitors of this protease domain and present the 2.35-Å structure of the CPD bound to one of these inhibitors. This structure, coupled with biochemical and mutational studies of the toxin, reveals the molecular basis of CPD substrate specificity and underscores the evolutionary relationship between the CPD and the clan CD caspase proteases. These studies are likely to prove valuable for devising new antitoxin strategies for a number of bacterial pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Satchell, K.J. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect. Immun. 75, 5079–5084 (2007).
Li, L., Rock, J.L. & Nelson, D.R. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76, 2620–2632 (2008).
Lee, B.C. et al. Vibrio vulnificus rtxE is important for virulence, and its expression is induced by exposure to host cells. Infect. Immun. 76, 1509–1517 (2008).
Lee, J.H. et al. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45, 146–152 (2007).
Liu, M., Alice, A.F., Naka, H. & Crosa, J.H. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 75, 3282–3289 (2007).
Olivier, V., Haines III, G.K., Tan, Y. & Satchell, K.J. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect. Immun. 75, 5035–5042 (2007).
Olivier, V., Salzman, N.H. & Satchell, K.J. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect. Immun. 75, 5043–5051 (2007).
Cordero, C.L., Sozhamannan, S. & Satchell, K.J. RTX toxin actin cross-linking activity in clinical and environmental isolates of Vibrio cholerae. J. Clin. Microbiol. 45, 2289–2292 (2007).
Rahman, M.H. et al. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 27, 347–355 (2008).
Sheahan, K.L., Cordero, C.L. & Satchell, K.J. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. USA 101, 9798–9803 (2004).
Sheahan, K.L. & Satchell, K.J. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell. Microbiol. 9, 1324–1335 (2007).
Sheahan, K.L., Cordero, C.L. & Satchell, K.J. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 26, 2552–2561 (2007).
Lupardus, P.J., Shen, A., Bogyo, M. & Garcia, K.C. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science 322, 265–268 (2008).
Egerer, M., Giesemann, T., Jank, T., Satchell, K.J. & Aktories, K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 282, 25314–25321 (2007).
Giesemann, T., Egerer, M., Jank, T. & Aktories, K. Processing of Clostridium difficile toxins. J. Med. Microbiol. 57, 690–696 (2008).
Reineke, J. et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446, 415–419 (2007).
Gordon, V.M. & Leppla, S.H. Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect. Immun. 62, 333–340 (1994).
Arastu-Kapur, S. et al. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 4, 203–213 (2008).
Powers, J.C., Asgian, J.L., Ekici, O.D. & James, K.E. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 102, 4639–4750 (2002).
Prochazkova, K. & Satchell, K.J. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J. Biol. Chem. 283, 23656–23664 (2008).
Asgian, J.L. et al. Aza-peptide epoxides: a new class of inhibitors selective for clan CD cysteine proteases. J. Med. Chem. 45, 4958–4960 (2002).
Ganesan, R. et al. Exploring the S4 and S1 prime subsite specificities in caspase-3 with aza-peptide epoxide inhibitors. Biochemistry 45, 9059–9067 (2006).
Barrett, A.J. & Rawlings, N.D. Evolutionary lines of cysteine peptidases. Biol. Chem. 382, 727–733 (2001).
Bedard, K.M. & Semler, B.L. Regulation of picornavirus gene expression. Microbes Infect. 6, 702–713 (2004).
Reed, K.E. & Rice, C.M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242, 55–84 (2000).
Schilling, O. & Overall, C.M. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 (2008).
Stennicke, H.R., Renatus, M., Meldal, M. & Salvesen, G.S. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 350, 563–568 (2000).
Eichinger, A. et al. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18, 5453–5462 (1999).
Kato, D. et al. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 1, 33–38 (2005).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, California, USA, 2002).
Acknowledgements
We thank P. Gulig (University of Florida) for generously providing Vibrio vulnificus CMCP6 genomic DNA, E. Shank and R. Kolter (Harvard Medical School) for providing the Photorhabdus luminescens TTO1 strain, and M. Blokech and G. Schoolnik (Stanford School of Medicine) for help with V. cholerae strain construction and for providing V. cholerae genomic DNA. P.J.L. is supported by the Damon Runyon Cancer Research Foundation. K.C.G. is supported by the Keck Foundation and the Howard Hughes Medical Institute. M.B. is supported by the Burroughs Wellcome Foundation, the Searle Scholars Program, the US National Institutes of Health National Technology Center for Networks and Pathways (grant U54-RR020843) and the Human Frontier Science Program (grant RGP0024/2006-C).
Author information
Authors and Affiliations
Contributions
The inhibitor screen, synthesis of AS01, AS02 and AS04, protein expression and purification, cleavage assays, FT-MS data analysis, V. cholerae strain construction, actin crosslinking assays, MARTXVc silver staining and western blot analyses were performed by A.S. Crystallization of the MARTX CPD–InsP6–JCP598 complex was performed by A.S. and P.J.L. P.J.L. collected the data, solved and analyzed the structure, and generated the figures of the inhibitor-bound CPD structure. J.C.P. provided the cysteine protease compound library. V.E.A. synthesized JCP598, AS01 and VEA223, guided A.S. in the synthesis of AS01, AS02 and AS04, and assessed the integrity of all compounds described in this paper. A.G. designed the conditions for running samples for FT-MS, ran the samples for FT-MS and provided advice in FT-MS analysis. Creative input and financial support for the project were provided by M.B. The manuscript was written by A.S. and M.B. with advice from P.J.L., V.E.A., K.C.G. and A.G.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 1 and Supplementary Methods (PDF 2078 kb)
Rights and permissions
About this article
Cite this article
Shen, A., Lupardus, P., Albrow, V. et al. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat Chem Biol 5, 469–478 (2009). https://doi.org/10.1038/nchembio.178
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.178
This article is cited by
-
C-terminal modification and functionalization of proteins via a self-cleavage tag triggered by a small molecule
Nature Communications (2023)
-
De novo design of modular and tunable protein biosensors
Nature (2021)
-
New ligation independent cloning vectors for expression of recombinant proteins with a self-cleaving CPD/6xHis-tag
BMC Biotechnology (2017)
-
Architecture of the type IV coupling protein complex of Legionella pneumophila
Nature Microbiology (2017)
-
Actin activates Pseudomonas aeruginosa ExoY nucleotidyl cyclase toxin and ExoY-like effector domains from MARTX toxins
Nature Communications (2016)