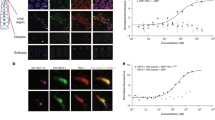
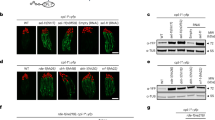
Abstract
Small RNAs direct RNA-induced silencing complexes (RISCs) to regulate stability and translation of mRNAs1,2. RISCs associated with target mRNAs often accumulate in discrete cytoplasmic foci known as GW-bodies3. However, RISC proteins can associate with membrane compartments such as the Golgi and endoplasmic reticulum4. Here, we show that GW-bodies are associated with late endosomes (multivesicular bodies, MVBs). Blocking the maturation of MVBs into lysosomes by loss of the tethering factor HPS4 (ref. 5) enhances short interfering RNA (siRNA)- and micro RNA (miRNA)-mediated silencing in Drosophila melanogaster and humans. It also triggers over-accumulation of GW-bodies. Blocking MVB formation by ESCRT (endosomal sorting complex required for transport)6 depletion results in impaired miRNA silencing and loss of GW-bodies. These results indicate that active RISCs are physically and functionally coupled to MVBs. We further show that MVBs promote the competence of RISCs in loading small RNAs. We suggest that the recycling of RISCs is promoted by MVBs, resulting in RISCs more effectively engaging with small RNA effectors and possibly target RNAs. It may provide a means to enhance the dynamics of RNA silencing in the cytoplasm.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
11 November 2009
In this letter, statements regarding the functions of HPS4 and 1 have been altered from those initially published, A section of text has been inserted, a sentence was altered and a sentence was deleted. This has been corrected in both the HTML and PDF versions of the article.
References
Zamore, P. D. & Haley, B. Ribo-gnome: the big world of small RNAs. Science 309, 1519–1524 (2005).
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 9, 102–114 (2008).
Cikaluk, D. E. et al. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell 10, 3357–3372 (1999).
Li, W. et al. Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays 26, 616–628 (2004).
Piper, R. C. & Katzmann, D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 (2007).
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004).
Vidal, M., Larson, D. E. & Cagan, R. L. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev. Cell 10, 33–44 (2006).
Lai, E. C., Tam, B. & Rubin, G. M. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 19, 1067–1080 (2005).
Wang, C. W., Stromhaug, P. E., Kauffman, E. J., Weisman, L. S. & Klionsky, D. J. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 163, 973–985 (2003).
Falcon-Perez, J. M., Nazarian, R., Sabatti, C. & Dell'Angelica, E. C. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell Sci. 118, 5243–5255 (2005).
Nguyen, T. & Wei, M. L. Hermansky-Pudlak HPS1/pale ear gene regulates epidermal and dermal melanocyte development. J. Invest. Dermatol. 127, 421–428 (2007).
Kinch, L. N. & Grishin, N. V. Longin-like folds identified in CHiPS and DUF254 proteins: vesicle trafficking complexes conserved in eukaryotic evolution. Protein Sci. 15, 2669–2674 (2006).
Williams, R. L. & Urbe, S. The emerging shape of the ESCRT machinery. Nature Rev. Mol. Cell Biol. 8, 355–368 (2007).
Lloyd, T. E. et al. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108, 261–269 (2002).
Vaccari, T. & Bilder, D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell 9, 687–698 (2005).
Miura, G. I., Roignant, J. Y., Wassef, M. & Treisman, J. E. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development 135, 1913–1922 (2008).
Behm-Ansmant, I. et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898 (2006).
Lin, M. D. et al. Drosophila processing bodies in oogenesis. Dev. Biol. 322, 276–288 (2008).
Pham, J. W., Pellino, J. L., Lee, Y. S., Carthew, R. W. & Sontheimer, E. J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117, 83–94 (2004).
Kim, K., Lee, Y. S. & Carthew, R. W. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13, 22–29 (2007).
Fielenbach, N. et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell 12, 443–455 (2007).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nature Rev. Mol. Cell Biol. 5, 739–751 (2004).
Bellare, P. et al. A role for ubiquitin in the spliceosome assembly pathway. Nature Struct. Mol. Biol. 15, 444–451 (2008).
Gibbings, D. J., Ciaudo, C. & Voinnet, O. Sorting of GW182 into multivesicular bodies controls microRNA activity. Nature Cell Biol. doi: 10:1038/ncb1930 (2009).
Kujala, P. et al. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75, 3873–3884 (2001).
Ding, S. W. & Voinnet, O. Antiviral immunity directed by small RNAs. Cell 130, 413–426 (2007).
Lugli, G., Larson, J., Martone, M. E., Jones, Y. & Smalheiser, N. R. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905 (2005).
Parton, R. G., Simons, K. & Dotti, C. G. Axonal and dendritic endocytic pathways in cultured neurons. J. Cell Biol. 119, 123–137 (1992).
Stoorvogel, W., Kleijmeer, M. J., Geuze, H. J. & Raposo, G. The biogenesis and functions of exosomes. Traffic 3, 321–330 (2002).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 9, 654–659 (2007).
Acknowledgements
We thank R. Cagan, H. Folsch, P. Sharp, G. J. Hannon, Q. Liu, X. Wang, C. Horvath, G. Goshima, V. Helfand, M. Lowe, I. Hariharan, D. Bilder, J. Treisman, M. Siomi and J. Pham for reagents; H. Folsch, I. Fields, A. Komuro, and D. Harris for help with some of the experiments; H. Jiang for help with statistics and the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for fly strains and antibodies. Y.S.L. was supported by a FRAXA postdoctoral Fellowship; J.J.C. was supported by the CMBD Training Grant and J.B.P. was supported by a Presidential Fellowship. This work was also supported by grants from the National Institutes of Health to E.J.S. (GM072830) and R.W.C. (GM77581, GM68743), and from a grant from the Korean BioGreen 21 Program to Y.S.L. (20070301034036).
Author information
Authors and Affiliations
Contributions
Y.L., E.S. and R.C. planned the project; all of the authors performed experimental work, except for E.S. and R.C.; Y.L., S.P., A.A., K.K., J.W., J.C., D.L., I.C., K.N., J.P., E.S. and R.C. performed data analysis and Y.L. and R.C wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare competing financial interests. R.W.C. and E.J.S. own shares in Silentech Inc., which may gain financially from publication. R.W.C. and Y.S.L. have a patent application whose value may be affected by publication.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1556 kb)
Rights and permissions
About this article
Cite this article
Lee, Y., Pressman, S., Andress, A. et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11, 1150–1156 (2009). https://doi.org/10.1038/ncb1930
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1930
This article is cited by
-
MicroRNA-mediated regulation of glucose and lipid metabolism
Nature Reviews Molecular Cell Biology (2021)
-
Extracellular RNA: mechanisms of it’s transporting into target cells
ExRNA (2019)
-
Urinary Biomarkers for Bladder Outlet Obstruction
Current Bladder Dysfunction Reports (2017)
-
Targeting microRNAs as key modulators of tumor immune response
Journal of Experimental & Clinical Cancer Research (2016)
-
Membrane-association of mRNA decapping factors is independent of stress in budding yeast
Scientific Reports (2016)