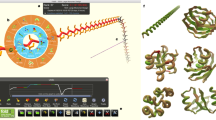
Abstract
People exert large amounts of problem-solving effort playing computer games. Simple image- and text-recognition tasks have been successfully ‘crowd-sourced’ through games1,2,3, but it is not clear if more complex scientific problems can be solved with human-directed computing. Protein structure prediction is one such problem: locating the biologically relevant native conformation of a protein is a formidable computational challenge given the very large size of the search space. Here we describe Foldit, a multiplayer online game that engages non-scientists in solving hard prediction problems. Foldit players interact with protein structures using direct manipulation tools and user-friendly versions of algorithms from the Rosetta structure prediction methodology4, while they compete and collaborate to optimize the computed energy. We show that top-ranked Foldit players excel at solving challenging structure refinement problems in which substantial backbone rearrangements are necessary to achieve the burial of hydrophobic residues. Players working collaboratively develop a rich assortment of new strategies and algorithms; unlike computational approaches, they explore not only the conformational space but also the space of possible search strategies. The integration of human visual problem-solving and strategy development capabilities with traditional computational algorithms through interactive multiplayer games is a powerful new approach to solving computationally-limited scientific problems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
von Ahn, L. & Dabbish, L. Labeling images with a computer game. in CHI '04: Proc. 2004 Conf. Human Factors Comput. Syst., 319–326 (ACM, 2004)
von Ahn, L., Liu, R. & Blum, M. Peekaboom: a game for locating objects in images. in CHI '06: Proc. SIGCHI Conf. Human Factors Comp. Syst., 55–64 (ACM, 2006)
Westphal, A. J. et al. Non-destructive search for interstellar dust using synchrotron microprobes. In X-ray Optics Microanalysis: Proc. 20th Int. Congr. Vol. 1221, 131–138 (2010)
Rohl, C., Strauss, C., Misura, K. & Baker, D. Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 (2004)
Anfinsen, C. B. Principles that govern the folding of protein chains. Science 181, 223–230 (1973)
Das, R. & Baker, D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 77, 363–382 (2008)
Qian, B. et al. High-resolution structure prediction and the crystallographic phase problem. Nature 450, 259–264 (2007)
Bradley, P., Misura, K. M. S. & Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871 (2005)
Yee, N. Motivations of play in online games. J. CyberPsychol. Behav. 9, 772–775 (2007)
Rosato, A. et al. CASD-NMR: critical assessment of automated structure determination by NMR. Nature Methods 6, 625–626 (2009)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002)
Acknowledgements
We thank D. Salesin, K. Tuite, J. Snyder, D. Suskin, P. Krähenbühl, A. C. Snyder, H. Lü, L. S. Tan, A. Chia, M. Yao, E. Butler, C. Carrico, P. Bradley, I. Davis, D. Kim, R. Das, W. Sheffler, J. Thompson, O. Lange, R. Vernon, B. Correia, D. Anderson, Y. Zhao, S. Herin and B. Bethurum for their help. We would like to thank N. Koga, R. Koga and A. Deacon and the JCSG for providing us with protein structures before their public release. We would also like to acknowledge all of the Foldit players who have made this work possible. Usernames of players whose solutions were used in figures can be found in Supplementary Table 4. This work was supported by NSF grants IIS0811902 and 0906026, DARPA grant N00173-08-1-G025, the DARPA PDP program, the Howard Hughes Medical Institute (D.B.), Microsoft, and an NVIDIA Fellowship. This material is based upon work supported by the National Science Foundation under a grant awarded in 2009.
Author information
Authors and Affiliations
Contributions
All named authors contributed extensively to development and analysis for the work presented in this paper. Foldit players (more than 57,000) contributed extensively through their feedback and gameplay, which generated the data for this paper.
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Figures S1-S14 with legends, Supplementary Tables S1-S4, Player Testimonials and References. (PDF 2788 kb)
Rights and permissions
About this article
Cite this article
Cooper, S., Khatib, F., Treuille, A. et al. Predicting protein structures with a multiplayer online game. Nature 466, 756–760 (2010). https://doi.org/10.1038/nature09304
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09304
This article is cited by
-
Leveraging the Potential of Large Language Models in Education Through Playful and Game-Based Learning
Educational Psychology Review (2024)
-
Machine culture
Nature Human Behaviour (2023)
-
Online citizen science with the Zooniverse for analysis of biological volumetric data
Histochemistry and Cell Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.