Abstract
Mental retardation (MR) is characterized by cognitive impairment with an IQ <70. Many of the major causes are genetically determined and the ∼30% male excess suggests that mutations in genes carried on the X chromosome are disproportionably represented. One such gene, jumonji AT-rich interactive domain 1C (JARID1C) on Xp11.2, has been identified in families with X-linked MR (XLMR), with 18 different mutations reported to date. As part of a systematic resequencing of 720 genes in 208 XLMR families of the International Genetic of Learning Disability (IGOLD) consortium, two novel nucleotide changes in the JARID1C coding region were identified, with the nucleotide changes segregating with the disease phenotype in the two families. The first mutation is a single-nucleotide insertion in exon 21 (c.3258_3259insC p.K1087fs*43) causing a frameshift and resulting in a premature termination codon (PTC). Such PTC-containing mRNAs are generally degraded by nonsense-mediated mRNA decay (NMD) surveillance, but our results show that this is not the case with this mutation. The other change is a single-nucleotide substitution in exon 12 (c.1160C>A) in a published family with nonsyndromic MR, MRX13. This change occurs in a highly conserved amino acid, with proline (P) being substituted by threonine (T) (p.P544T). Functional analysis shows that this amino-acid substitution compromises both tri- and didemethylase activity of the JARID1C protein. We conclude that the two novel changes impair JARID1C protein function and are disease-causing mutations in these families.
Similar content being viewed by others
Introduction
The jumonji, A/T-rich interactive domain 1C (JARID1C, also termed SMCX) gene is located on the short arm of the X chromosome at Xp11.22-p11.21. The gene encodes a 1560 amino-acid protein that belongs to the JARID1 subfamily of Arid DNA-binding proteins.1 The JARID1C protein contains several conserved DNA motifs, such as a JmjN domain, a JmjC domain, an ARID/BRIGHT DNA-binding domain, a C5HC2 zinc-finger domain and PHD zinc-finger domains.2, 3 The protein possesses H3K4me3-specific demethylase activity and is shown to function as a transcriptional repressor through the RE-1-silencing transcription factor (REST) complex.4, 5
Mutations in the JARID1C gene have been reported in patients with X-linked mental retardation (XLMR).2, 6, 7, 8, 9 Eighteen mutations in JARID1C have been reported to date and are spread throughout the gene. Affected individuals with JARID1C mutations show a mild-to-severe range of intellectual disability. There is also a report of a mutation in a patient with autism spectrum disorder.7
In this study, we identified two novel nucleotide changes in the coding region of JARID1C in two XLMR families. One of these families is an unreported XLMR family, whereas the other is the MRX13 family.10 The first change is a single-nucleotide insertion (c.3258_3259insC), which is predicted to cause a frameshift and subsequently a premature termination codon (PTC). The other change, in the MRX13 family, is a single-nucleotide substitution in exon 12 (c.1160C>A). This change occurs at a highly conserved amino acid in the JmjC domain of JARID1C (p.P544T). Our functional assay shows that this amino-acid substitution compromises both JARID1C H3K4 tri- and didemethylase activity. This is the first study showing functional consequence of a naturally occurring missense mutation in JARID1C in a family with nonsyndromic MR.
Materials and methods
XLMR families
Family 1
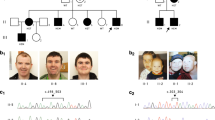
This family is of southeast European descent and consists of three affected brothers (Figure 1a) first examined in 1996. Their medical records documented that all three had severe intellectual disability with speech delay. All three were recorded to have head circumferences within the normal range, with heights on the third percentile, and all three were documented to be epileptic, with individual IV-1 noted to have absence seizures.
Family 1. (a) Pedigree of family 1 and DNA sequencing result of the proband compared with that of a normal control. The pedigree shows that the mutation, c.3258_3259insC, arose as a de novo event in individual III-1 and is detected in all three affected individuals. The DNA sequencing result shows the C nucleotide insertion as compared with that of a normal male control. (b) Picture of the proband from family 1 showing his face and bulbous fingertips.
The most recent examination of the proband (IV-2) was conducted at the age of 25 years. He lives in a group home and has severe intellectual impairment. His mother was unsure of his early motor milestones but remembered that he was slow to talk. He speaks only a single word (‘No’) but uses signs quite effectively and has some basic comprehension. His general health is good. On examination, his head circumference was 56 cm (50th centile). His height and weight were not recorded but he had obvious short stature. He tended to hold his head down and avoided eye contact. He has prominent ears, nose and eyebrows (Figure 1b). Fundal examination was normal. Teeth were uneven with a normally shaped palate. A freckled pigmented area was present on the right side of his neck. He had flexion contractures of about 30° at his elbows and had bulbous fingertips, broad feet and slightly bulbous great toes. His tone was generally increased but variable. Reflexes were brisk and symmetrical. His right plantar response was downgoing and the left was upgoing.
From observation it is probable that the proband's mother (III-1) had learning difficulties and her mother (II-1) was also suspected to have some degree of intellectual disability as she was unable to read or write. However this may partly be explained by the fact that she did not speak or understand English when she migrated to Australia.
Family 2
This family was ascertained in 1985 and published as nonsyndromic XLMR family MRX13 (Figure 2).10 The disease locus was linked to markers in the Xp22.3–Xq21.22 region.10 More recent clinical reviews of individuals III-6 and III-7 showed moderate intellectual disability, short stature and microcephaly with heights of 165 and 163 cm (3rd percentile), and both with head circumferences of 53 cm (3rd centile). Facially, they were nondysmorphic, apart from both men having high nasal bridges and large ears measuring 7 cm (>98th centile). Both have pleasant personalities. Of the six female carriers within the family, four have normal intelligence, one (III-8) has some learning difficulties but has managed to maintain a household and rear a family, and one (IV-13) has a mild-to-moderate intellectual disability and works in sheltered employment. Their physical examinations were unremarkable. Individual IV-17 has a mild intellectual disability but was not a carrier. She had a ventricular septal defect (VSD), normal height (157 cm) and microcephaly with a head circumference of 51 cm (<3rd centile) and tapering fingers. She had a son who died at birth from multiple congenital anomalies (cleft lip, unilateral microtia, diaphragmatic hernia and VSD). Whole-genome array CGH did not reveal any microdeletion or duplication in either IV-10 or her son.
Family 2 (MRX13). (a) Pedigree of the family. Three affected individuals and six female carriers have been identified. (b) DNA sequencing result of the proband shows a C>A nucleotide substitution as compared with that of a normal male control. (c) The amino-acid alignment shows that the proline at position 544, p.P544, is highly conserved across orthologs and known paralogs in the JARID1 family.
Major clinical features of affected males from both families are summarized in Table 1.
Mutation analysis
As part of the International Genetic of Learning Disability (IGOLD) consortium, we performed a systematic resequencing of 720 genes in 208 XLMR families.11 We identified two unique nucleotide changes in the coding region of the JARID1C gene in families 1 and 2.
Segregation analysis was performed in both families. In family 1, exon 21 was amplified using forward primer (5′-CACTGGTGAGTGGACCCT-3′) and reverse primer (5′-CCCCTTAGCTGTTACCGTCA-3′). In family 2, exon 12 was amplified using forward primer (5′-CAGTAGATGCCGGTGTTGTG-3′) and reverse primer (5′-GAGAGGGAATGACTGGGCTT-3′). All PCRs were carried out on 100 ng of genomic DNA from each individual, with 1 × PCR (Roche, Indianapolis, IN, USA), 1.5 mM MgCl2, 200 μ M each of dATP, dCTP, dGTP and dTTP, 0.5 μ M each of forward and reverse primers and 1 U of Taq polymerase (Roche). The PCRs were processed through an initial denaturing step at 94°C for 5 min, then through 35 amplification cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s and final extension at 72°C for 5 min. The PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA) and were sequenced using a BigDye Terminator V.3.1 cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
RT-PCR of the JARID1C c.3258_3259insC mutated allele
Total RNA was extracted from the lymphoblastoid cell line (LCL) of the proband from family 1 using a Qiagen RNeasy Mini Kit (Qiagen). cDNA was generated using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) from 2 μg of total RNA. cDNA of 2 μl (1:10 of the cDNA reaction) was used in a PCR containing 1 × PCR (Roche), 1.5 mM MgCl2, 200 μ M each of dATP, dCTP, dGTP and dTTP, 0.5 μ M each of forward primer in exon 20 (5′-TGCCCAACATCCAGGCTCTC-3′), reverse primer in exon 23 (5′-GATACCCTCCTTCTCCTTCTG-3′) and 1 U of Taq polymerase (Roche). The PCRs were processed through an initial denaturing step at 94°C for 5 min, then through 35 amplification cycles of 94°C for 30 s, 61.5°C for 30 s, 72°C for 30 s and final extension at 72°C for 5 min. The expected PCR products of 437 bp in size were visualized by electrophoresis on a 1.5% agarose gel after staining with ethidium bromide. The PCR products were subsequently purified using a QIAquick PCR Purification Kit (Qiagen). DNA sequencing was performed on both strands using a BigDye Terminator V.3.1 cycle Sequencing Kit (Applied Biosystems).
Site-directed in vitro mutagenesis of the c.1160C>A mutation
In vitro mutagenesis was performed on a full-length JARID1C gene4 in the Gateway Entry System (Invitrogen) using a QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with the mutagenesis primer (5′-GGCTGGCTATCAAATAGTTCAGTTGTCAGCTTCTTCATCACTTC-3′) to introduce c.1160C>A mutation. The procedure was performed according to the manufacturer's protocol. Nucleotide sequencing of the entire coding sequence of the construct was performed to confirm the change.
Functional analysis of the p.P544T protein
Functional analysis was performed by expressing mutated protein in insect Sf9 cells.4 The protein was purified and testing for demethylation activity was carried out using histone peptides representing di- and trimethylation of K4 on H3.4 The result was measured by MALDI-TOF mass spectrometry as described in Iwase et al.4 Comparison between wild-type and mutated p.P544T proteins was performed. The relative activity was calculated from the ratio of the demethylated peptide to the input of the uncatalyzed peptide.4
Results
Systematic resequencing of the coding exons of 720 genes on the X chromosome in probands from 208 families with XLMR identified two novel nucleotide changes in the coding region of the JARID1C gene in two families. In the first family, composed of three affected brothers IV-1, IV-2 and IV-4, a single-nucleotide insertion in exon 21 (c.3258_3259insC) was detected in all three and their mother (III-1), an obligate carrier (Figure 1a). The insertion is not present in the maternal grandmother (II-1), indicating that it arose de novo in III-1. The insertion is predicted to cause a frameshift, and thus result in the creation of a PTC 43-amino acids downstream. Such PTC-containing mRNAs are usually degraded by nonsense-mediated mRNA decay (NMD) surveillance; hence, semiquantitative RT-PCR was performed on total RNA extracted from the proband's LCL and three normal controls. The result shows that the mRNA transcript containing the insertion was detected in similar quantities as in the controls and was therefore unlikely to be subjected to NMD (Figure 3). In addition, no abnormally sized RT-PCR product was detected, suggesting that this insertion is also unlikely to lead to alternative splicing of JARID1C mRNA. Taken together, and supported by published data, we believe that this c.3258_3259insC insertion represents a novel, deleterious JARID1C mutation.
Gel electrophoresis result of the RT-PCR of the JARID1C gene from the proband in family 1 (P) with three normal controls (C1, C2 and C3). The mutated transcript containing c.3258_3259insC is present, as also shown by DNA sequencing. Thus, mRNA from the mutated allele seems to be insensitive or only partially sensitive to NMD.
In the MRX13 family proband, IV-3, we found a single-nucleotide substitution in exon 12 (c.1160C>A), which was also identified in the two other affected males in the family. This nucleotide change results in the substitution of proline with threonine at position 544 (p.P544T). Proline 544 lies within the core of the JmjC domain of JARID1C and is highly conserved among JARID1C orthologs and its paralogs (Figure 2c). The JARID1C gene lies within the linkage interval of the MRX13 family. Individual IV-17 does not have the mutation. Given that her and her son's phenotypes differ from other family members, it is likely that there is a different etiology for her problems.
The JARID1C protein was previously shown to function as a specific H3K4me3 and H3K4me2 demethylase.4 This demethylase catalytic activity of JARID1C resides within the JmjC domain. To determine whether the p.P544T change affects the demethylase activity of JARID1C, mutated and wild-type proteins were expressed in and purified from insect Sf9 cells. The p.P544T mutant protein was similarly soluble and stable when compared with wild-type protein (Figure 4a). The outcome of the demethylation reaction was measured by MALDI-TOF spectrometry. The results showed that the p.P544T missense change led to a significantly reduced demethylase activity of JARID1C (Figure 4b). Thus, these results suggest that the c.1160C>A, p.P544T change is a disease-causing mutation, and that reduced demethylase activity of the JARID1C protein is the underlying pathology of XLMR in the MRX13 family.
Histone demethylation activity of the mutated JARID1C protein (p.P544T) as compared with that of a normal control. (a) Both p.P544T and wild-type JARID1C proteins were expressed in insect Sf9 cells and both seemed to be stable and of expected size. (b) Di- and trimethylated synthetic histone peptides were incubated with wild-type JARID1C and mutated JARID1C (p.P544T) protein. Demethylated peptides were detected as peaks indicated by arrows. The mutated JARID1C protein (p.P544T) is shown to decrease the histone demethylase activity for H3K4me2 and H3K4me3 when compared with the wild-type protein.
Discussion
In this study, we identified two novel mutations in the JARID1C gene.
In the first family, a c.3258_3259insC, p.K1087fs*43 mutation has been identified. This change most likely arose de novo in individual III-1. The possibility that it had been inherited from the maternal grandfather (II-2) is much less likely, given the severity of the JARID1C phenotype and the fact that males with intellectual disability are much less likely to reproduce. Incomplete penetrance might offer another explanation; however, this has not yet been formally reported for JARID1C mutations. This nucleotide insertion is predicted to cause a frameshift and thus a truncated protein. The resulting PTC-containing mRNA is not degraded by NMD, most likely because of the fact that the PTC, which is created as a consequence of the c.3258_3259insC insertion, is only 52 nucleotides from the exon 22–23 boundary. Messenger RNAs containing PTC codons 50–55 bp upstream of the exon–exon junction, or 25–30 nucleotides upstream of the exon junction complex, are often insensitive or partially sensitive to NMD.12 The truncated protein will lack the C-terminal end and as such the second PHD zinc-finger domain (Figure 5). The C-terminus of the JARID1C protein was previously shown to be crucial for protein localization.5 Mislocalization of the protein, most likely into the cytoplasm, will interrupt the protein function as a transcriptional repressor. In addition, PHD domains have been shown to be histone–methyl–lysine-binding motifs13, 14 and the first PHD domain was reported to bind H3K9me3 preferentially.4 Lack of one PHD domain will likely compromise the cross talk between JARID1C and other methylated histones, resulting in an impaired gene function.
The second mutation, c.1160C>A, p.P544T, is located in the JmjC domain, which is responsible for the histone demethylase enzymatic activity of JARID1C.5 Together with another missense change recently published,6 these are the only two mutations predicted to affect the Jmjc domain function directly. Both mutations have been found in families with relatively mild intellectual disability. In this study, we showed that a missense change in the Jmjc domain of JARID1C has the potential to impair its histone demethylase activity. However, the JmjC domain is not solely responsible for JARID1C demethylase activity. The JmjN domain, located at the N-terminus, was also shown to contribute to the enzymatic activity of the protein. Deletion of the JmjN domain can abolish demethylase activity as well.3, 5 As such, the phenotypic effect of any missense mutation is predicted to be less severe.
JARID1C mutations and their phenotypes
Mutations in the JARID1C gene generate a spectrum of phenotypes, from nonsyndromic MR (NS-MR) to syndromic MR. The most common features observed in affected individuals reported to date are hyperreflexia, short stature, aggressive behavior and seizures.6 Microcephaly has been reported infrequently.6, 7, 9 The JARID1C protein was previously identified with REST at the RE1 elements of several target genes.5 Some of them are neuronal genes associated with epilepsy and psychiatric disorders, such as SCN2A, CACNAH1 and SLC6A3.15, 16, 17, 18, 19, 20 Interruption of gene function may interfere with those pathways, thus causing epileptic and psychiatric phenotypes.
Although there is no apparent specific phenotype for individuals with JARID1C mutations, severity of the phenotype may depend on the type and position of the mutation. Currently, there are 20 mutations, including the two reported in this study, in 52 affected males (Figure 5, Table 2).2, 6, 7, 8, 9 Some carrier females, with generally mild intellectual disability, have also been reported. These include three carrier females from the K8545 family of Abidi et al,6 one carrier female from the family N063 of Jensen et al,2 one from family A015 of Tzschach et al,9 and one from Family 1 and two from Family 2 (see above) reported here. Altogether, there were eight generally mildly affected females. However, in at least some of these pedigrees, familial aggregation of intellectual disability, independent of JARID1C, was also reported (eg, individual IV-17 in Family 2, see above; or the discordant female carrier monozygotic twins in family K8545 of Abidi et al6). Whether the female manifestation is because of the respective JARID1C mutation expression is suggestive, but not conclusive. Small numbers preclude us from drawing any meaningful conclusions.
Nonsense and frameshift mutations
Three nonsense mutations have been reported. The mRNA transcripts from affected alleles in two families were confirmed to undergo NMD.2 There are two insertion mutations and one single-nucleotide deletion that have been previously reported. All cause a frameshift and truncation of the JARID1C protein.21 The affected individuals with either nonsense or frameshift mutations tend to have severe intellectual disability. Most of them, where documented, also have short stature. Four out of six probands also had either seizures or abnormal EEGs.2, 6, 9 Considering that the C-terminus of the protein is important in the targeting of the JARID1C protein to the nucleus,5 such mutations can be considered as complete loss of function mutations and thus represent the most severe spectrum of clinical presentations. There is also one report of a point mutation in intron 11 resulting in skipping of exon 11 and, as a consequence, a frameshift and a PTC-containing JARID1C mRNA.6 The affected individuals with this mutation presented with severe intellectual disability and mild dysmorphic features, but no seizures.
Missense mutations
Thirteen missense mutations in the JARID1C gene, spanning the coding region, have been reported so far.2, 6, 7, 9 The severity of the associated phenotype varies. Most mutations cluster in and around the C5HC2 zinc-finger domain. The majority of the mutations located in the C5HC2 domain and between JmjC and C5HC2 result in severe MR with or without epilepsy (Figure 5, Table 2). The function of the C5HC2 domain is still unclear. The domain itself contains eight potential zinc ligand-binding residues and may have a DNA-binding function.22 The C5HC2 domain is also conserved among proteins in the JARID1 family. Functional studies of four mutations located in this region (p.F642L, p.L731F, p.E698K and p.Y751C)4, 5 showed that all nucleotide substitutions cause partial loss of function of histone demethylase. Proteins harboring mutations that cause less-severe phenotypes (p.Y751C) tend to have higher levels of enzymatic activity.4, 5 On the basis of these observations, it is reasonable to speculate that the C5HC2 domain may also have a role in the demethylation of the downstream targeted genes. Thus, nucleotide changes in this region are likely to have a significant impact on overall gene function.
In conclusion, we identified two novel mutations in the coding region of the JARID1C gene. The nucleotide insertion, c.3258_3259insC, generates a frameshift resulting in a PTC. This mutation causes severe intellectual impairment and epilepsy with mild dysmorphic features. The second mutation is a nucleotide substitution, c.1160C>A, p.P544T, in the JmjC domain. This mutation causes a partial loss of the H3K4 histone demethylase activity of JARID1C. Affected individuals have moderate intellectual impairment, short stature and microcephaly. Our results in conjunction with other published reports indicate that mutations in JARID1C are an important cause of XLMR and that a more defined genotype–phenotype correlation is emerging.
References
Kortschak RD, Tucker PW, Saint R : ARID proteins come in from the desert. Trends Biochem Sci 2000; 25: 294–299.
Jensen LR, Amende M, Gurok U et al: Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 2005; 76: 227–236.
Kim TD, Shin S, Janknecht R : Repression of Smad3 activity by histone demethylase SMCX/JARID1C. Biochem Biophys Res Commun 2008; 366: 563–567.
Iwase S, Lan F, Bayliss P et al: The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007; 128: 1077–1088.
Tahiliani M, Mei P, Fang R et al: The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007; 447: 601–605.
Abidi FE, Holloway L, Moore CA et al: Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J Med Genet 2008; 45: 787–793.
Adegbola A, Gao H, Sommer S, Browning M : A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am J Med Genet A 2008; 146A: 505–511.
Santos C, Rodriguez-Revenga L, Madrigal I, Badenas C, Pineda M, Mila M : A novel mutation in JARID1C gene associated with mental retardation. Eur J Hum Genet 2006; 14: 583–586.
Tzschach A, Lenzner S, Moser B et al: Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat 2006; 27: 389.
Kerr B, Gedeon A, Mulley J, Turner G : Localization of non-specific X-linked mental retardation genes. Am J Med Genet 1992; 43: 392–401.
Tarpey P, Smith R, Pleasance E et al: A systematic, large-scale resequencing screen of the X chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543.
Isken O, Maquat LE : The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 2008; 9: 669–712.
Shi X, Hong T, Walter KL et al: ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006; 442: 96–99.
Wysocka J, Swigut T, Xiao H et al: A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006; 442: 86–90.
Berkovic SF, Heron SE, Giordano L et al: Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Ann Neurol 2004; 55: 550–557.
Cook Jr EH, Stein MA, Krasowski MD et al: Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet 1995; 56: 993–998.
Heron SE, Phillips HA, Mulley JC et al: Genetic variation of CACNA1H in idiopathic generalized epilepsy. Ann Neurol 2004; 55: 595–596.
Heron SE, Khosravani H, Varela D et al: Extended spectrum of idiopathic generalized epilepsies associated with CACNA1H functional variants. Ann Neurol 2007; 62: 560–568.
Nelson KB, Grether JK, Croen LA et al: Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann Neurol 2001; 49: 597–606.
Weiss LA, Escayg A, Kearney JA et al: Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry 2003; 8: 186–194.
Delbridge ML, Longepied G, Depetris D et al: TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X-implications for Y chromosome evolution. Chromosome Res 2004; 12: 345–356.
Yi L, Hao Z, Yang T, Wang S, Xing B, Xu Y : cDNA cloning, bioinformatic and tissue-specific expression analysis of porcine JARID1C gene. J Genet Genomics 2007; 34: 1088–1096.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rujirabanjerd, S., Nelson, J., Tarpey, P. et al. Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype–phenotype correlation. Eur J Hum Genet 18, 330–335 (2010). https://doi.org/10.1038/ejhg.2009.175
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.175
Keywords
This article is cited by
-
Expanding the genetics and phenotypic spectrum of Lysine-specific demethylase 5C (KDM5C): a report of 13 novel variants
European Journal of Human Genetics (2023)
-
A female case with novel KDM5C heterozygous variation presenting with Claes-Jensen type-like phonotype
BMC Neurology (2022)
-
Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C
Clinical Epigenetics (2018)
-
Spermatogenic failure and the Y chromosome
Human Genetics (2017)
-
Xp11.2 microduplications including IQSEC2, TSPYL2 and KDM5C genes in patients with neurodevelopmental disorders
European Journal of Human Genetics (2016)