Abstract
Genome-wide association studies (GWAS) using a large number of single nucleotide polymorphisms (SNPs) have successfully been applied to identify genetic variants of common diseases. However, genotyping using the new array technologies is often associated with spurious results that could unfavorably affect analyses of GWAS. Consequently, data cleaning is of paramount importance in excluding spurious genotyping results. In this study, we investigated the criteria required for the appropriate cleaning of 389 unrelated healthy Japanese samples analyzed using the GeneChip Human Mapping 500K Array Set for GWAS. The samples were randomly subdivided into two groups, and the allele frequencies in the groups were compared for individual SNPs as a quasi-case-control study. Then, observed results were filtered by four parameters (SNP call rate, confidence score obtained using the Bayesian Robust Linear Model with Mahalanobis genotype-calling algorithm, Hardy–Weinberg equilibrium, and minor allele frequency) and assessed for deviation from the null hypothesis. We found that appropriate data cleaning could be achieved using these four parameters. Our findings offer an avenue for obtaining appropriate data from GWAS.
Similar content being viewed by others
Introduction
One of the goals underlying the study of common diseases is to identify susceptibility and/or resistance genes associated with them. Previous studies of common diseases include two broad categories: family-based linkage studies across the entire genome, and population-based case-control association studies of candidate genes. Although there have been notable successes, progress has been slow. Linkage studies usually have low power except when a single locus explains a substantial fraction of the disease. In case-control association studies of candidate genes, researchers can target only those genes that have been functionally described.
In contrast, new high-throughput single nucleotide polymorphism (SNP) typing technologies for genome-wide association studies (GWAS) have recently been launched (Matsuzaki et al. 2004; Oliphant et al. 2002). GWAS provide opportunities to identify novel susceptibility and/or resistance loci without any prior information about gene functions. GWAS have successfully identified genetic variants associated with common diseases, including macular degeneration, QT interval prolongation, Crohn’s disease, type 2 diabetes, and cerebral infarction (Arking et al. 2006; Dewan et al. 2006; Klein et al. 2005; Kubo et al. 2007; Rioux et al. 2007; Sladek et al. 2007; The Wellcome Trust Case Control Consortium 2007).
However, one fundamental problem is that the results obtained by the new array technologies are not always accurate. In such large data sets, small systematic differences can produce effects that are capable of obscuring the true associations being sought (Clayton et al. 2005; Zondervan and Cardon 2004). The major causes of inaccurate results are systematic errors of array reaction, incomplete genotype-calling algorithms, and SNPs in regions that exhibit copy number variations. Less accurate genotyping data unfavorably affects GWAS analyses. Therefore, data cleaning is of paramount importance, and data should be checked thoroughly (Balding 2006). At present, a consensus for data-cleaning criteria has not been established. In this study, we assessed the following parameters for data cleaning: (1) SNP call rate, (2) confidence score in the Bayesian Robust Linear Model with Mahalanobis (BRLMM) genotype-calling algorithm, (3) fitness to Hardy–Weinberg equilibrium (HWE), and (4) minor allele frequency (MAF). We typed 389 unrelated healthy Japanese samples by the GeneChip Human Mapping 500K Array Set and analyzed different thresholds for these four parameters.
Materials and methods
Subjects and genotyping
The subjects included 389 unrelated, healthy Japanese individuals living in Japan. This study was approved by the research ethics review committees of the University of Tokyo.
Genotyping of 500,568 SNPs was performed using the GeneChip Human Mapping 500K Array Set (Affymetrix). This array set consisted of two chips (Sty I and Nsp I) with approximately 250,000 SNPs each that were used for each individual. Approximately 250 ng of genomic DNA was digested with two restriction enzymes (StyI and NspI) and processed according to the manufacturer’s protocol. The genotyping calls were analyzed using the GCOS 1.4 and GTYPE 4.1 software packages, which adopted the BRLMM genotype-calling algorithm (Rabbee and Speed 2006). BRLMM performs a multiple-chip analysis, enabling simultaneous estimation of probe effects and allele signals for each SNP. The confidence score in BRLMM is assigned for each observation according to the normalized distance from the center of the genotype cluster. The confidence score is d1/d2, where d1 is the smallest distance of the three and d2 is the second-smallest distance. The confidence score threshold is the maximum score at which the algorithm will make a genotype call. All lower-quality confidence calls with scores greater than the threshold result in a no call.
Assessment of data cleaning
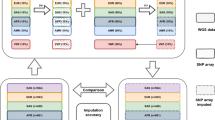
To assess the appropriate data-cleaning methods, the 389 healthy control samples were divided into two temporary groups (group A and group B). Each group was separately analyzed by the BRLMM algorithm of GTYPE 4.1 because the analytical capacity of this software was 250 samples (maximum). Then, to remove the bias associated with BRLMM analysis, group A and B samples were equally subdivided into two new groups (group 1 and group 2); group 1 consisted half of group A’s and half of group B’s samples, and group 2 consisted the remaining samples (Fig. 1). As a quasi-case-control study, group 1 was compared with group 2 using the chi-square test for the difference between allele frequencies of each SNP. SNPs on chromosome X were omitted because these groups were not matched by gender. Next, for reshuffling analysis, 100 new combination sets were prepared using the same 389 healthy control samples. To confirm that the appropriate data cleaning was reproducible in sets having the bias associated with BRLMM analysis, each set was formed from two groups separately analyzed by the BRLMM algorithm, such as the set of groups A and B (Fig. 1). Each set was also analyzed by the chi-square test for the difference in allele frequencies of each SNP.
The principle for making groups. The 389 healthy control samples were divided into two temporary groups (groups A and B). Each group was separately analyzed by the Bayesian Robust Linear Model with Mahalanobis (BRLMM) of GTYPE 4.1 software. Then, to remove the bias associated with the BRLMM analysis, the samples of groups A and B were equally subdivided into two new groups (groups 1 and 2); group 1 consisted half of group A’ s and half of group B’ s samples, and group 2 consisted of the remaining samples. Group 1 was compared with group 2 using the chi-square test for the difference between allele frequencies of each single nucleotide polymorphism. Next, for reshuffling analysis, 100 new combination sets were prepared using the same 389 healthy control samples. Each set was formed from two groups separately analyzed by the BRLMM algorithm, such as the set of groups A and B
The following four parameters were assessed in this surveillance:
-
1.
SNP call rate: to remove an SNP for which genotyping was consistently problematic, SNPs with call rates ≥85, ≥90 and ≥95% in both groups were prepared.
-
2.
Confidence score with the BRLMM: Confidence scores of 0.3, 0.4, 0.5, and 0.6 were applied. Each overall call rate was 93.3% (StyI: 92.2%; NspI: 94.4%) for a confidence score of 0.3; 95.3% (StyI: 94.4%; NspI: 96.1%) for 0.4; 96.6% (StyI: 96.0%; NspI: 97.2%) for 0.5; and 97.6% (StyI: 97.1%; NspI: 98.0%) for 0.6 before data cleaning. For a confidence score of 0.5, the distribution of call rates (per SNP and sample) is shown in Supplementary Figs. 1 and 2.
-
3.
HWE: Deviations from HWE can occur by chance. However, they can also be due to genotyping errors, inbreeding, and population stratification. Testing for HWE can be helpful to check data (Balding 2006). We removed SNPs for which we observed genotype frequencies that significantly deviated from HWE (HWE P < 0.001 and P < 0.01). Evaluation of HWE was carried out using the chi-square test. In general, case-control studies, SNPs that deviate from HWE in a control group are removed. We considered one group in a set as controls. The possibility that a deviation from HWE is due to a deletion or duplication polymorphism, which could be important for disease susceptibility, should now be considered (Bailey and Eichler 2006; Conrad et al. 2006; Nielsen et al. 1998).
-
4.
MAF: We removed SNPs in which the minor allele frequency was <1 or <5% in all samples.
SNPs with low MAF would produce inappropriately small P values in the chi-square test. However, in this study, the chi-square test could be used for the quasi-case-control studies and evaluation of HWE because SNPs with low MAF were removed for data cleaning.
First, the number of significant SNPs in a quasi-case-control study (P < 0.0001 and P < 0.001) was counted for each result after data cleaning and compared against the expected number calculated from each P value and the total number of SNPs. Next, the log quantile–quantile (QQ) P value (Balding 2006; Weir et al. 2004) was adopted for interpreting each result. The negative logarithm of P values was plotted against –log (i/(L + 1)), where L is the number of SNPs. Deviation from the expected number and the y = x line corresponds to loci that deviate from the null hypothesis. The close adherence of P values to the expected number and the expected line, which corresponds to the null hypothesis over most of the range, is encouraging, as it implies that there are few systematic sources of spurious association because mutual healthy control groups were compared.
Results
Criteria for data cleaning
We compared two healthy groups (groups 1 and 2, Fig. 1) using the chi-square test as a quasi-case-control study. Then, we assessed the deviation of the results from the null hypothesis after each data cleaning (see “Materials and methods” for details). A small or no deviation implies that there are few systematic sources of spurious associations.
-
1.
SNP call rate
As shown in Table 1, the number of SNPs with a call rate ≥95% was close to the expected number calculated from each P value and the total number of SNPs. For a confidence score of 0.5, the ratio of observed and expected number of SNPs with a call rate ≥95% was 1.05–1.50, whereas the ratio with call rates of ≥90% and ≥85% was 1.35–2.47 and 1.52–2.96, respectively (Table 1). The observed number of significant SNPs with call rates ≥90% and ≥85% was more inflated. For other confidence scores (0.3, 0.4, and 0.6), the ratio of observed and expected number of SNPs with a call rate ≥95% was 1.05–2.04, whereas the ratio with call rates ≥90% and ≥85% was 1.08–3.39 and 1.21–3.73, respectively (Supplementary Table 1). SNPs with call rates ≥90% and ≥85% also caused inflations. However, for a confidence score of 0.5, the ratio of observed and expected number of SNPs with a call rate ≥95% was up to 1.50. Nine significant SNPs with P < 0.0001 in the same region (within about 100 kb) were responsible for this random deviation. These SNPs, showing uniformly low P values, existed in a linkage disequilibrium block with a solid spine of D′ > 0.8. It was shown that SNPs with a lower call rate are likely to contain genotyping errors, and SNP call rate is important for data cleaning.
Next, thresholds of SNP call rate ranging from 92% to 97% at increments of 1% were set, and the ratio of observed and expected number of SNPs in each threshold was calculated (Fig. 2). This ratio was saturated at SNPs with a call rate ≥95%, implying that they had few spurious associations and were considered to be the key threshold. We focused on SNPs with a call rate ≥95% in the following analyses using other parameters.
Ratio of observed and expected number of single nucleotide polymorphisms (SNPs) with a call rate at 1% intervals between 92% and 97%. Ratio of observed and expected number of SNPs was calculated in each call rate, a confidence score of 0.5, Hardy–Weinberg equilibrium (HWE) P ≥ 0.001 and minor allele frequency (MAF) ≥5%
-
2.
Confidence score in BRLMM
In the BRLMM analysis, the ratio of observed and expected number of SNPs was 1.05–1.56 for confidence scores of 0.3, 0.4, and 0.5. However, this ranged from 1.13 to 2.04 for a confidence score of 0.6 (Table 2). Consequently, a confidence score of 0.6 would cause spurious associations. The total number of SNPs for a confidence score of 0.5 ranged from 229,907 to 262,854, whereas for confidence scores of 0.3 and 0.4, this was 146,469–217,818 (Table 2). On equivalent adequacy, the total number of SNPs for each confidence score (0.3, 0.4, and 0.5) should be taken into consideration. Then, the overall call rate for a confidence score was 93.3% for 0.3, 95.3% for 0.4, and 96.6% for 0.5. Therefore, it is suggested that a confidence score of 0.5 should be selected.
-
3.
HWE
SNPs with an HWE of P ≥ 0.001 or P ≥ 0.01 did not result in unexpected inflations. For a confidence score of 0.5, the ratio of observed and expected number of SNPs with an HWE of P ≥ 0.001 was 1.05–1.50, whereas for HWE with P ≥ 0.01, it was 1.05–1.48 (Table 1). We can conclude that, as hundreds of thousands of SNPs were analyzed in this study, deviation from HWE might be caused by chance for SNPs with an HWE of P ≥ 0.001.
-
4.
MAF
SNPs with MAF ≥5% or ≥1% did not exhibit unexpected inflations. For a confidence score of 0.5, the ratio of observed and expected number of SNPs for MAF ≥5% was 1.10–1.50, whereas for MAF ≥1%, this was 1.05–1.48 (Table 1). The decision regarding which SNPs with MAF ≥5% or ≥1% should be adopted depends on the sample size of each association study.
The log QQ P value plot was described after data cleaning using the criteria identified by the above analyses (SNP call rate ≥95%, confidence score 0.5, HWE P ≥ 0.001, and MAF ≥5% or ≥1%; Fig. 3a, b). Plots of P values were close to the expected line (y = x). However, approximately 12 SNPs deviated from the expected line of low P values. The nine significant SNPs with P < 0.0001 in a linkage disequilibrium block with a solid spine of D′ > 0.8 were also responsible for this random deviation.
Log quantile–quantile (QQ) P value plot for the results after data cleaning. a and b were analyzed using groups 1 and 2, respectively. Cleaning criteria were a single nucleotide polymorphism (SNP) call rate ≥95%, confidence score 0.5, Hardy–Weinberg equilibrium (HWE) P ≥ 0.001, and minor allele frequency (MAF) ≥5%; b SNP call rate ≥95%, confidence score 0.5, HWE P ≥ 0.001, and MAF ≥1%
Taken together, these results indicated that data cleaning could be appropriately conducted in our Japanese samples using SNP call rate ≥95%, a confidence score of 0.5, HWE P ≥ 0.001, and MAF ≥5% or ≥1%.
Reshuffling analysis
Next, to confirm that the identified appropriate criteria were reproducible, 100 new combination sets were prepared. In addition, to consider that the bias associated with BRLMM analysis cannot be removed when other investigators make use of our frequency data, each set was formed from two groups separately analyzed by the BRLMM algorithm, such as the set of groups A and B (Fig. 1). Then, two groups in each set were compared as a quasi-case-control study. The mean ratio of observed and expected number of SNPs with a call rate of ≥95% was 0.91–0.98, whereas the mean ratio with call rates of ≥90% and ≥85% was 1.21–3.62 and 2.74–18.18, respectively (Table 3). Unexpected deviations were not observed in comparisons of these 100 sets when data cleaning was conducted using SNP call rate ≥95%, a confidence score of 0.5, HWE P ≥ 0.001, and MAF ≥5 or ≥1%.
Discussion
GWAS have considerable potential for detecting susceptibility and/or resistant genes for various complex diseases. However, the considerable amount of data may occasionally create difficulties for investigators. One hundred thousand to 1 million SNPs are targeted for GWAS. It is important to note that the results obtained from genotyping so many SNPs are not always precise and that inaccurate data will unfavorably affect GWAS analyses. In this study, it was shown that spurious associations could be excluded using the criteria that we identified. However, it may not be possible to apply these criteria to data from the GeneChip Human Mapping 500K Array Set or other arrays used by other investigators, because the criteria may be affected by differences in overall call rates between studies. In such cases, the same analyses using four parameters (SNP call rate, confidence score in BRLMM, HWE, and MAF) would facilitate identification of appropriate criteria for each study. In the GeneChip Human Mapping 500K Array Set, these criteria can be applied to data that is of the same quality as our data, with an index for overall call rate (for a confidence score of 0.5, overall call rate was 96.6% before data cleaning in this study).
The tradeoff exists between overall call rate and accuracy. If a high accuracy is of greater importance than a high overall call rate, a higher-quality threshold for data cleaning should be selected. Alternatively, if a high overall call rate is of greater importance than a high accuracy, a lower quality threshold for data cleaning should be selected. In this study, we assessed each data-cleaning method by the deviation from the null hypothesis to obtain accurate data. Additionally, on equivalent adequacy, a higher overall call rate was considered. Therefore, the appropriate data cleaning methods we identified satisfy both overall call rate and accuracy.
In GWAS with the GeneChip Human Mapping 500K Array Set, the genotyping results of case–control samples were decided by the BRLMM algorithm. In the reshuffling analysis (100 sets), two groups in each set were separately analyzed by the BRLMM algorithm. The maximum mean ratio of observed and expected number in the reshuffling analysis was up to 18.18 (Table 2), whereas that in groups 1 and 2, equally subdivided from groups A and B, was 2.95 (Table 1). This result suggested that separate BRLMM analyses result in a bias of genotyping results. However, as shown in Table 3, unexpected deviations were not observed in the reshuffling analysis after the appropriate data cleaning (cleaning criteria: SNP call rate ≥95%, a confidence score of 0.5, HWE P ≥ 0.001, and MAF ≥5 or ≥1%). We assume that the appropriate data cleaning could remove SNPs affected by the bias associated with BRLMM analysis.
Some studies required that samples pass a threshold of overall call rate (Buch et al. 2007; Rioux et al. 2007; The Wellcome Trust Case Control Consortium 2007), whereas we evaluated data cleaning methods for SNP selection. As a result, the data cleaning methods we identified could exclude spurious associations, suggesting that it would not be necessary to require sample selection when the appropriate data cleaning for SNP selection is conducted.
The four parameters (SNP call rate, confidence score in BRLMM, HWE, and MAF) are mutually correlated. In these four parameters, SNP call rate was considered to be a key parameter because SNPs with a lower call rate, particularly, caused irrelevant inflations (Tables 1, 3; Fig. 2). In GWAS, it would be better to change the threshold of SNP call rate when the ratio of observed and expected number of SNPs with low P values is inflated. If the ratio is close to one according to the threshold of SNP call rate, it is suspected that SNPs with low P values include spurious associations due to errors as well as true associations with the target disease.
In the reshuffling analysis, we calculated deviations from expected P values of each chip (StyI and NspI). Before data cleaning, the maximum mean ratio of observed and expected number of SNPs on StyI chip was 47.08, whereas that of SNPs on NspI chip was 30.10 (Supplementary Table 2). Thus, the mean ratio for StyI chip was more inflated. There might be a difference in the accuracy between the two chips before data cleaning. However, after the appropriate data cleaning, the maximum mean ratio of observed and expected number of SNPs on StyI chip was 0.98, whereas that of SNPs on NspI chip was 0.93 (Supplementary Table 2). Accordingly, unexpected deviations were not observed in either chip after the appropriate data cleaning.
Even though data cleaning using appropriate criteria was conducted, sample size should be carefully considered when SNPs with an MAF ≥1% are used. The frequency of an SNP has a marked influence on statistical power. To identify SNPs with low MAF associated with complex disease, sample size must be large (Ohashi and Tokunaga 2001, 2002; Ohashi et al. 2001; Risch 2000). In instances where SNPs with low MAF are used, at the very least, desirable sample sizes should be calculated based on the frequencies of the targeted SNPs, and sufficient samples should be collected when planning the GWAS.
Generally, 300,000 SNPs might be required to capture most of the common genetic variation in a population (Balding 2006). The GeneChip Human Mapping 500K Array Set provides genotyping data for approximately 500,000 SNPs. However, only about 250,000 SNPs were extracted after data cleaning in our Japanese samples (Table 1). This reduction is caused by the presence of numerous SNPs with low MAF on the GeneChip Human Mapping 500K Array Set in Japanese. There are approximately 150,000 SNPs with an MAF <5% and 100,000 SNPs with an MAF <1% on this array set. Ideally, SNPs with low MAF are not likely to be suitable for GWAS (Ohashi and Tokunaga 2001, 2002; Ohashi et al. 2001; Risch 2000), and it is hoped that new arrays for different ethnic groups will be developed.
References
Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A (2006) A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet 38:644–651
Bailey JA, Eichler EE (2006) Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 7:552–564
Balding DJ (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7:781–791
Buch S, Schafmayer C, Volzke H, Becker C, Franke A, von Eller-Eberstein H, Kluck C, Bassmann I, Brosch M, Lammert F, Miquel JF, Nervi F, Wittig M, Rosskopf D, Timm B, Holl C, Seeger M, ElSharawy A, Lu T, Egberts J, Fandrich F, Folsch UR, Krawczak M, Schreiber S, Nurnberg P, Tepel J, Hampe J (2007) A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet 39:995–999
Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, Nutland S, Howson JM, Faham M, Moorhead M, Jones HB, Falkowski M, Hardenbol P, Willis TD, Todd JA (2005) Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet 37:1243–1246
Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK (2006) A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38:75–81
Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J (2006) HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314:989–992
Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389
Kubo M, Hata J, Ninomiya T, Matsuda K, Yonemoto K, Nakano T, Matsushita T, Yamazaki K, Ohnishi Y, Saito S, Kitazono T, Ibayashi S, Sueishi K, Iida M, Nakamura Y, Kiyohara Y (2007) A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nat Genet 39:212–217
Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R (2004) Parallel genotyping of over 10, 000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res 14:414–425
Nielsen DM, Ehm MG, Weir BS (1998) Detecting marker-disease association by testing for Hardy–Weinberg disequilibrium at a marker locus. Am J Hum Genet 63:1531–1540
Ohashi J, Tokunaga K (2001) The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J Hum Genet 46:478–482
Ohashi J, Tokunaga K (2002) The expected power of genome-wide linkage disequilibrium testing using single nucleotide polymorphism markers for detecting a low-frequency disease variant. Ann Hum Genet 66:297–306
Ohashi J, Yamamoto S, Tsuchiya N, Hatta Y, Komata T, Matsushita M, Tokunaga K (2001) Comparison of statistical power between 2 × 2 allele frequency and allele positivity tables in case-control studies of complex disease genes. Ann Hum Genet 65:197–206
Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl:56–58, 60–61
Rabbee N, Speed TP (2006) A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics 22:7–12
Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR (2007) Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39:596–604
Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856
Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885
The Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678
Weir BS, Hill WG, Cardon LR (2004) Allelic association patterns for a dense SNP map. Genet Epidemiol 27:442–450
Zondervan KT, Cardon LR (2004) The complex interplay among factors that influence allelic association. Nat Rev Genet 5:89–100
Acknowledgments
We are deeply grateful to the people participating in this study. We thank JAMSAC (Japan Multiple System Atrophy Research Consortium) for a part of the samples. This study is supported by a grant-in-aid for Scientific Research on Priority Areas “Comprehensive Genomics” and “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan and a grant-in-aid for JSPS fellows.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyagawa, T., Nishida, N., Ohashi, J. et al. Appropriate data cleaning methods for genome-wide association study. J Hum Genet 53, 886–893 (2008). https://doi.org/10.1007/s10038-008-0322-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0322-y
Keywords
This article is cited by
-
Understanding Mendelian errors in SNP arrays data using a Gochu Asturcelta pig pedigree: genomic alterations, family size and calling errors
Scientific Reports (2022)
-
Genome-wide association study of idiopathic hypersomnia in a Japanese population
Sleep and Biological Rhythms (2022)
-
Identification of hybridization and introgression between Cinnamomum kanehirae Hayata and C. camphora (L.) Presl using genotyping-by-sequencing
Scientific Reports (2020)
-
Adaptive selection of founder segments and epistatic control of plant height in the MAGIC winter wheat population WM-800
BMC Genomics (2018)
-
Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis
Scientific Reports (2016)