Abstract
Expression of the cellular inhibitor of apoptosis protein 1 (cIAP1) is unexpectedly repressed at the level of translation under normal physiological conditions in many cell lines. We have previously shown that the 5′ untranslated region of cIAP1 mRNA contains a stress-inducible internal ribosome entry site (IRES) that governs expression of cIAP1 protein. Although inactive in unstressed cells, the IRES supports cap-independent translation of cIAP1 in response to endoplasmic reticulum stress. To gain an insight into the mechanism of cIAP1 IRES function, we empirically derived the minimal free energy secondary structure of the cIAP1 IRES using enzymatic cleavage mapping. We subsequently used RNA affinity chromatography to identify several cellular proteins, including nuclear factor 45 (NF45) as cIAP1 IRES binding proteins. In this report we show that NF45 is a novel RNA binding protein that enhances IRES-dependent translation of endogenous cIAP1. Further, we show that NF45 is required for IRES-mediated induction of cIAP1 protein during the unfolded protein response. The data presented are consistent with a model in which translation of cIAP1 is governed, at least in part, by NF45, a novel cellular IRES trans-acting factor.
Similar content being viewed by others
Main
Eukaryotes have evolved distinct mechanisms that allow them to control the expression of their proteome independent of the transcriptional apparatus. The ability to regulate translation ensures rapid and measured expression of specific proteins in time and space. Translational control allows for efficient reprogramming of gene expression during cell growth and differentiation1, 2 and provides a level of homeostatic control following exposure to stresses such as viral infection,3 endoplasmic reticulum (ER) stress,4, 5 hypoxia6 or DNA damage.7 Regulation of translation occurs primarily at the initiation step and targets several eukaryotic initiation factors that mediate recruitment of the ribosome to the mRNA before polypeptide synthesis.8
The 7-methyl-guanosine cap located at the 5′ end of mature mRNAs catalyzes the formation of a protein complex consisting of a cap binding protein (eIF4E), a scaffold protein (eIF4G) and an RNA helicase (eIF4A). Together, these proteins comprise the cap binding complex (eIF4F), which allows for the recruitment of the small ribosomal subunit (along with accessory factors) forming a preinitiation complex that is believed to proceed in a 5′–3′ direction along the 5′ untranslated region (UTR) until an optimal initiation codon is reached. It is at this point that the large ribosomal subunit joins to form the 80S ribosome and peptide synthesis commences.8 This cap-dependent, ribosomal scanning mechanism of translation initiation works efficiently under normal physiological conditions; however, during times of cellular stress, decreased availability of ATP and the preinitiation complex significantly reduces overall protein synthesis rates.
To respond properly to various physiological stimuli, cells must be able to ensure expression of specific genes despite repressed global translation rates. One such mechanism uses an RNA sequence element located in the 5′ UTR that facilitates recruitment of the ribosome. First discovered in picornaviruses, the internal ribosome entry site (IRES) element is present in a number of eukaryotic mRNAs where it mediates cap-independent translation initiation under stress conditions.9 The exact mechanism used by eukaryotic IRES elements to recruit the ribosome is the subject of intense investigation. Unlike some viral IRES, which can function by direct recruitment of the ribosome, eukaryotic IRES appears to require accessory protein factors in addition to canonical initiation factors. Several of these IRES trans-acting factors (ITAFs) have been identified, including PTB,10 hnRNPA1,11 La12 and hnRNPC1/C2.13 Exactly how ITAFs function in modulating cellular IRES activity is not clear. ITAFs were suggested to function as adapter proteins acting as a bridge between the ribosome and RNA.14 Alternatively, they may exert their effect as RNA chaperones, remodeling RNA into a conformation that is permissive to ribosome recruitment.15
Cellular inhibitor of apoptosis protein 1 (cIAP1) is a critical regulator of cell survival and nuclear factor-κB (NF-κB) signaling.16, 17 We and others have shown that the expression of cIAP1 is regulated at the level of translation through an IRES located within its 5′ UTR that supports cap-independent translation.5, 18, 19 Although this IRES is inactive in unstressed cells, drug-induced ER stress that leads to the unfolded protein response (UPR), DNA damage by etoposide treatment or cell-cycle arrest by sodium arsenite treatment causes an increase in cIAP1 IRES activity. Importantly, the concomitant increase in cIAP1 protein levels during the UPR delays the onset of apoptosis, consistent with the antiapoptotic role of cIAP1.
To better understand the regulation and function of cIAP1 IRES, we derived the minimum free energy secondary structure of the cIAP1 IRES using in vitro enzymatic cleavage mapping. Furthermore, we identified a specific cohort of cIAP1 IRES binding proteins including nuclear factor 45 (NF45). NF45 was first identified as an NFAT-related transcription factor that together with its binding partner NF90 regulates interleukin-2 transcription.20 Here we ascribe a novel, post-transcriptional role for NF45. Specifically, we found that NF45 enhances IRES-mediated translation of cIAP1 mRNA. Surprisingly, we found that NF45 alone possesses RNA binding activity, interacts specifically with the cIAP1 IRES in vitro and modulates cIAP1 IRES activity in vivo. More importantly, cells lacking NF45 failed to upregulate IRES-mediated translation of cIAP1 during the UPR. Our data show that NF45 is a novel RNA binding protein that interacts with the cIAP1 5′ UTR in a sequence and structure dependent manner and regulates expression of cIAP1 in response to stress.
Results
Minimum free energy secondary structure model of the cIAP1 IRES
We have mapped the cIAP1 IRES activity to nucleotides −150 to −80 (relative to AUG start codon) of the 1.2-kb-long cIAP1 5′ UTR.18 In addition to the primary sequence, secondary and tertiary structures are important determinants of cellular IRES activity.21 To determine whether the cIAP1 IRES shares common structural features with other cellular IRES and to better understand the role that RNA structure may have in the regulation of the cIAP1 IRES activity, we empirically determined the secondary structure of the cIAP1 IRES by primer extension analysis of nuclease-digested RNA fragments. The cIAP1 IRES was in vitro transcribed and subjected to digestion with RNase T1, T2 or V1. cDNA was then generated from the resulting RNA fragments by reverse transcription and resolved on a denaturing polyacrylamide gel (Supplementary Figure S1). Using the digest pattern as folding constraints for the mfold structure prediction algorithm,22 we obtained a minimum free energy model of the cIAP1 IRES structure (Figure 1a; Supplementary Table S1).
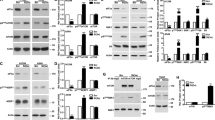
cIAP1 IRES structure model and identification of binding proteins. (a) The cIAP1 IRES RNA secondary structure model (nucleotides −150 to −1) with sites sensitive to RNase T1, T2 and V1 shown as shaded circles. (b) SYPRO Ruby-stained RNA affinity chromatography gel of proteins pulled down with a biotin-tagged cIAP1 IRES RNA probe (probe 1) from an HEK293T cytoplasmic lysate. (c) Peptides identified from excised bands by mass spectrometry were mapped to four unique proteins. (d) To confirm protein binding specificity, RNA affinity chromatography was repeated with a cIAP1 IRES (probe 1) or non-IRES (probe 2) RNA, followed by immunoblotting with antibodies specific for RHA, NF90, IGF2BP1 and NF45
A specific set of proteins interacts with the cIAP1 IRES
A cursory examination of the highly structured cIAP1 IRES reveals a partial likeness to the IRES of cricket paralysis virus (CrPV). Stem-loops I, II and III of the cIAP1 IRES (Figure 1a) bear a striking similarity to pseudoknots II and III of the CrPV IRES – motifs that mediate direct interaction with the 40S ribosomal subunit.23 We therefore wished to determine if cIAP1 IRES RNA is capable of a direct interaction with the 40S ribosomal subunit. Although CrPV IRES was able to bind directly to the purified 40S in an electromobility shift assay we did not observe any binding with the cIAP1 IRES (data not shown). This suggests that additional cellular proteins are required for the recruitment of the cIAP1 IRES to the ribosome. To identify these proteins, we carried out RNA electromobility shift and UV cross-linking assays using a cytoplasmic extract from HEK293T cells and in vitro transcribed and radiolabeled RNA corresponding to the cIAP1 IRES. We observed specific protein–RNA complexes forming under both native (Supplementary Figure S2a) and denaturing (Supplementary Figure S2b) conditions, supporting the notion that specific proteins may regulate cIAP1 IRES activity.
To resolve and identify these proteins we used an RNA affinity chromatography strategy followed by identification of RNA-bound proteins by MALDI-TOF mass spectrometry. RNA encoding the cIAP1 IRES (nucleotides −150 to −1) or the non-IRES portion of the cIAP1 5′ UTR (nucleotides −80 to −1) was in vitro transcribed, 5′ end-labeled with biotin and conjugated to streptavidin-coated agarose beads. The labeled RNA was then incubated with a cytoplasmic HEK293T cell lysate and bound proteins were resolved by SDS-PAGE and visualized with SYPRO Ruby stain. A specific set of proteins was observed binding to the cIAP1 IRES but not in a control reaction lacking an RNA matrix (Figure 1b). Proteins resolved by affinity chromatography were excised from the gel and submitted for analysis by MALDI-TOF mass spectrometry. Peptides were mapped with high confidence to four distinct proteins using the MASCOT database (Figure 1c). Specifically, we found that RNA Helicase A (RHA; UniProt accession: Q08211), insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1; UniProt accession: Q9NZI8), NF90 (UniProt accession: Q12906) and NF45 (UniProt accession: Q12905) interacted with the cIAP1 IRES.
To confirm that these proteins interact specifically with the cIAP1 IRES, we repeated the affinity chromatography experiment and transferred the bound proteins to a membrane followed by immunoblotting with antibodies specific for RHA, NF90, IGF2BP1 and NF45. Importantly, these four proteins were confirmed to interact with the cIAP1 IRES but not to the portion of the cIAP1 5′ UTR that does not exhibit IRES activity (probe 2), indicating that these interactions are specific to the cIAP1 IRES sequence and/or structure (Figure 1d). Furthermore, probing the affinity preparations with antibodies specific for the canonical RNA binding proteins hnRNPA1, HuR and TIA1 did not yield a positive signal, showing that only a subset of RNA binding proteins interact with the cIAP1 IRES (Lewis et al.11; data not shown).
NF45 enhances translation of endogenous cIAP1
We chose to focus our investigation of cIAP1 IRES binding proteins on NF45, as relatively little is known about this protein's function in translation. NF45 was initially implicated, along with its binding partner NF90, as an interleukin-2 transcription factor in Jurkat T cells.20 Complicating the study of NF45 function is its apparent co-regulation with NF90. A recent study by Guan et al.24 and our own unpublished observations showed that removing NF45 triggers proteasomal degradation of NF90 and vice versa. The intimate relationship between NF45 and NF90 also extends to their apparent function in modulating protein translation, as the NF45–NF90 complex has been shown by Merrill et al.25 to bind and inhibit human rhinovirus 2 (HRV2) IRES activity. Consistent with their finding, we found that U373MG glioblastoma cells express approximately 50% less NF45 and NF90 protein relative to HEK293T cells (Figure 2a). Curiously, we found that U373 MG and HEK293T cells express similar levels of cIAP1 protein as determined by western blot (Figure 2a). However, when we assessed levels of newly synthesized cIAP1 protein by metabolic labeling and immunoprecipitation of cIAP1, we observed that translation of cIAP1 was reduced in U373 MG cells relative to HEK293T cells (Figure 2b). We observed a similar reduction in cIAP1 translation after transiently knocking down NF45 in HEK293T cells using siRNA (Supplementary Figure S3). Of note, although the inhibitory NF45/NF90 heterodimers are present in high abundance in HEK293T cells, both the abundance and the activity of this heterodimer are significantly lower in cell lines of glial origin.26 Therefore, we chose U373 MG glioblastoma cells as a model to address the function of NF45 in an NF90-independent manner.
NF45 regulates cIAP1 mRNA translation. (a) Western blots illustrating differential expression of endogenous NF45 and NF90 in HEK293T and U373 MG cells. (b) Metabolic labeling and immunoprecipitation of cIAP1 shows impaired translation of cIAP1 in U373 MG relative to HEK293T cells. (c) NF45 increases de novo cIAP1 protein synthesis. Top panel: U373 MG cells transiently expressing NF45 or GFP were pulse-labeled 24 h following transfection and newly synthesized cIAP1 and β-actin proteins were co-immunoprecipitated, resolved on SDS-PAGE and detected by autoradiography. Bottom panel: western blot of U373 MG cells transiently transfected with a FLAG-tagged NF45 overexpression plasmid illustrating efficient expression of the transgene. (d) NF45 enhances cIAP1 protein translation but not transcription. Top panel: densitometric analysis of cIAP1 protein expression from experiments performed in panel c (*n=2, P<0.001; mean±S.D.). Bottom Panel: quantitative RT-PCR of total cIAP1 mRNA levels in cells expressing GFP or NF45 (**n=3, P=0.33; mean±S.D.). (e) NF45 enhances translation efficiency of cIAP1 mRNA as measured by polysome profiling of U373 MG cells transiently expressing GFP or NF45. The efficiency of the separation was assessed by resolving ethidium-bromide-stained 28S and 18S rRNA from each fraction. Individual fractions were probed for cIAP1 or β-actin (control) mRNA expression by RT-PCR. PCR products were resolved by gel electrophoresis and densitometry was performed to determine the percent distribution of specific mRNAs across the gradient (n=3; mean±S.D.). The amount of specific mRNA present in higher-order polysomes (P) relative to the translationally quiescent pool (M) produces the metric P/M, and quantifies the change in translational efficiency
We transiently transfected U373 MG glioblastoma cells with a FLAG-tagged NF45 overexpression plasmid. Western blot analysis indicated a 1.5- to 2-fold enhancement of NF45 expression in transfected cells (Figure 2c, lower panels). To determine the effect of NF45 on de novo cIAP1 protein translation, U373 MG cells overexpressing NF45 or GFP as a control were pulse-labeled with 35S-methionine for 25 min, followed by immunoprecipitation of cIAP1 and β-actin. Although the global translation rate did not appear to be significantly affected by NF45 overexpression, cIAP1 translation was enhanced approximately 2.5-fold relative to β-actin translation (Figure 2c and densitometric analysis in upper panel of d). NF45 was originally described in the literature as a transcription factor. Therefore, it was plausible that our observations could be the result of NF45 targeting cIAP1 at the transcriptional level. We therefore assessed the steady-state levels of cIAP1 mRNA in U373 MG cells transiently transfected with NF45 overexpression plasmid by quantitative RT-PCR. Despite efficient expression of the NF45 relative to GFP-transfected cells (Figure 2c), no significant change was observed in cIAP1 mRNA levels (Figure 2d, lower panel).
To confirm that NF45 indeed affects the efficiency of cIAP1 mRNA translation, we assessed the polysomal distribution of cIAP1 mRNA in cells overexpressing GFP or NF45. A quantitative shift (1.5-fold increase in the polysome/monosome ratio of cIAP1 versus β-actin) into the higher-order polysome fractions was observed in NF45 overexpressing cells whereas no significant shift was observed in the β-actin mRNA pools, confirming that NF45 specifically enhances translation of cIAP1 mRNA (Figure 2e). Together, these data support a model in which NF45 enhances translation of endogenous cIAP1 mRNA.
NF45-dependent translation of cIAP1 is mediated by its IRES
We next sought to determine the mechanism by which NF45 alters the translational efficiency of cIAP1 mRNA. We initially identified NF45 interacting with the portion of the cIAP1 5′ UTR that confers IRES activity; therefore we hypothesized that NF45 modulates cIAP1 IRES activity. We have previously established a bicistronic reporter system to test the IRES activity in which the cIAP1 5′ UTR is inserted downstream of a β-galactosidase (β-GAL) cistron and upstream of a chloramphenicol transferase (CAT) cistron.27 The ratio of CAT expression to β-GAL activity yields a relative measure of IRES activity. The bicistronic vector was transiently co-transfected with a plasmid expressing full-length NF45 or GFP into U373MG cells. Cells transiently expressing NF45 exhibited a 240% increase in cIAP1 IRES activity relative to cells co-transfected with GFP (Figure 3a). Notably, this increase in cIAP1 IRES activity closely correlated with the increase in endogenous cIAP1 protein translation that we observed in our metabolic labeling/immunoprecipitation experiment (≈230% relative to GFP-expressing cells; Figure 2d, upper panel).
NF45 regulates IRES-dependent cIAP1 translation. (a) U373 MG cells were assayed 24 h following transient transfection of a bicistronic DNA reporter construct (schematic) containing the cIAP1 5′ UTR. IRES activity is expressed as the ratio of CAT expression over β-GAL activity. (b) U373 MG cells were transiently transfected with GFP or NF45 together with a monocistronic CAT reporter plasmid with either the cIAP1 5′ UTR (1.) or a truncated UTR that does not bind NF45 (2.) inserted upstream of the CAT reporter. A β-GAL reporter was co-transfected to normalize CAT expression across samples. (*P<0.05, **P<0.005, n=3 mean±S.D.)
To confirm that NF45 acts in a UTR-specific manner, we compared a monocistronic expression vector in which the 5′ UTR of cIAP1 (nucleotides −1222 to −1) or the portion of the 5′ UTR to which NF45 does not bind (nucleotides −80 to −1) is inserted upstream of the CAT cistron. DNA transfection was normalized by co-transfection with a β-GAL expression plasmid. We observed a significant increase in translation of the downstream CAT reporter in NF45-overexpressing cells with the full-length 5′ UTR (Figure 3b, construct 1) but not the IRES-deleted 5′ UTR (Figure 3b, construct 2).
We next asked whether the ITAF activity of NF45 is specific to the cIAP1 IRES or whether it could modulate other cellular IRES. To address this question, we assessed the IRES activity of cIAP1, APAF1,28 BclxL,29 DAP530 and VCIP31 in HeLa cell lines (HeLa cells express high levels of NF45 comparable to HEK293T) that stably express shRNA directed against NF45 (d5 cells) or shRNA with no specific target (c cells). This NF45 knockdown cell line has been previously characterized.24 As expected, we found that cIAP1 IRES activity was significantly reduced in cells expressing low amounts of NF45 (d5 versus c cells) (Figure 4a). Further, these data suggest that the mechanism of NF45-mediated regulation of cIAP1 IRES activity is conserved in glioblastoma and HeLa cells lineages. Importantly, we also found that there was no significant effect on either BclxL or APAF1 IRES activity, whereas we observed a small but consistent increase in DAP5 and VCIP IRES activity in the absence of NF45 (Figure 4a). These data confirm that NF45 functions to enhance translation in a UTR-dependent manner, specifically using the cIAP1 IRES element.
NF45 is required for IRES-mediated translation of cIAP1 during ER stress. (a) Knockdown of NF45 specifically impairs cIAP1 IRES activity. IRES activity in d5 (NF45 shRNA) cells relative to C (nontargeting shRNA) cells was measured using a bicistronic assay for cIAP1, APAF1, BclxL, DAP5 and VCIP as detailed in Material and methods (n=3, mean±S.D.). (b) NF45 is required for ER stress-mediated induction of cIAP1. c or d5 cells were treated with DMSO (D) as a vehicle or 5 μM thapsigargin (T) to induce ER stress for 24 h. Western blots were performed with antibodies against the indicated proteins. (c) Rescue of d5 cells with transient overexpression of NF45 but not NF90 enhances cIAP1 expression. d5 cells were transiently transfected with an OMNI-tagged NF90 construct (O-NF90c) or a FLAG-tagged NF45 construct harboring a silent mutation that renders it resistant to NF45 shRNA (F.NF45r). Cells were then treated with DMSO (D) or 5 μM thapsigargin (T) for 24 h. Western blots were performed with antibodies against the indicated proteins. Blots shown are representative of at least three experiments
Translation of cIAP1 during the unfolded protein response requires NF45
Induction of cIAP1 following ER stress has been observed in our laboratory and others. We have previously shown that IRES-mediated translation of cIAP1 is enhanced following ER stress and that p86, the ER stress-induced cleavage product of p97/DAP5, can enhance cIAP1 IRES activity.18 However, we were unable to observe direct binding of p86 to the cIAP1 IRES. This could be due to the transient interaction of p86 with the IRES, or an indirect interaction requiring an intermediary protein. Therefore, we asked whether NF45 could mediate ER stress induction of cIAP1 downstream of p86. To answer this question we again used the d5 cell line that expresses low levels of NF45. We have previously shown that induction of ER stress with the calcium ATPase inhibitor thapsigargin leads to increased cIAP1 protein through enhanced IRES activity in HeLa cells.18 We treated HeLa cell lines stably expressing control shRNA (c) or shRNA targeting NF45 (d5) with DMSO (D) vehicle or thapsigargin (T), for 24 h and assessed steady-state levels of cIAP1 by western blot (Figure 4b). As expected, we observed an increase in cIAP1 protein levels relative to GAPDH in control cells. However, no such increase was observed in the cell line lacking NF45 (d5) despite the fact that these cells show no impairment in the UPR pathway as shown by the induction of the protein chaperone BiP.
The lack of NF45 in d5 cells also results in decreased stability of its in vivo binding partner, NF90.24 To determine whether NF45 by itself is sufficient to mediate an increase in cIAP1 translation or whether the phenomenon also requires NF90, we performed a rescue experiment by transiently transfecting d5 cells before treatment with thapsigargin with either (1) an overexpression plasmid coding for an epitope-tagged NF90 (O.NF90c) or (2) an epitope-tagged version of NF45 that is resistant to NF45 shRNA (F.NF45r). We observed that despite increased levels of exogenous NF90 in transfected d5 cells (Figure 4c) there was still no induction of cIAP1 after ER stress. In contrast, cells expressing shRNA-resistant NF45 exhibited an increase in endogenous cIAP1 expression, whereas there was no observable additive effect of both thapsigargin treatment and NF45 overexpression on cIAP1 expression. These data show that NF45 is physiologically relevant and that it is sufficient to regulate cIAP1 levels during the UPR.
NF45 is a novel RNA binding protein
NF45 possesses no experimentally verified RNA binding activity and circumstantial evidence points to NF45 interacting with RNA indirectly through NF90. Recent data suggest that NF45 is able to bind directly to dsDNA, specifically to purine-rich boxes within the SP-10 and interleukin-2 promoters.32 As the affinity chromatography experiment may not distinguish between direct and indirect protein–RNA interactions, we attempted to photocross-link Escherichia coli expressed and purified, full-length NF45 (Figure 5a) to the cIAP1 5′ UTR in vitro. Recombinant GST-NF45, but not GST alone directly interacted with the portion of the cIAP1 5′ UTR that exhibits IRES activity (Figure 5b, probe 3). Moreover, the NF45–RNA complex was not efficiently formed on an RNA probe corresponding to the non-IRES portion of the 5′ UTR (Figure 5b, probe 2), an observation that is consistent with the results of the affinity chromatography experiments (Figure 1d). As a portion of probes 2 and 3 overlap (nucleotides −80 to −63, comprising SLII), we can conclude that sequence and/or structure common to this region participate in the formation of the NF45–RNA complex.
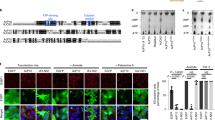
NF45 interacts directly with the cIAP1 IRES. (a) Recombinant GST-NF45 used for in vitro binding studies was expressed in E. coli using IPTG, affinity purified and assessed by SDS-PAGE as shown. (b) Recombinant NF45 (300 ng) was UV cross-linked to a 32P-labeled cIAP1 IRES RNA probe (nucleotides −150 to −63, probe 3) or a portion of the 5′ UTR with no IRES activity (nucleotides −80 to −1, probe 2). The RNA–protein complex was resolved by SDS-PAGE and detected by autoradiography. (c) Determination of NF45 binding sites on the cIAP1 IRES using masking DNA oligonucleotides. Experiments were performed as in b using RNA probe 3, except that the probe was first hybridized with each of eight competition DNA oligonucleotides (100-fold molar excess). The top panel shows a primary structure of the probe with the location of the masking oligos and the bottom panel illustrates oligo locations on the cIAP1 IRES secondary structure. (d) Confirmation of NF45 binding sites on the cIAP1 IRES by mutational analysis. Experiments were performed as in b with probe 3 (3wild type) or a mutant probe 3 with the bulge of SLI removed (3ΔCUUA). Alternatively, a probe corresponding to SLI of the cIAP1 IRES (SLIwild type) was used together with a sequence mutant that preserves the stem-loop structure (SLIs), or a sequence mutant that also disrupts base pairing at the base of SLI (SLIss)
To further delineate the NF45 binding site within the cIAP1 5′ UTR, we designed competitive DNA oligonucleotides spanning nucleotides −150 to −63 and hybridized them to a heat-denatured RNA probe before renaturation and incubation with recombinant NF45 to compete with the potential binding sites. Figure 5c shows that oligos 1, 2 and 3 successfully compete out NF45 binding whereas smaller oligos 2a/b and 3b fail to compete with binding. We observed partial competition with oligos 3a and 4. On the basis of these observations and analysis of the primary structure, we hypothesized that the base of stem-loop I is essential for direct interaction with NF45 (refer to SLI in Figure 1a). Specifically, the structure of stem-loop I is likely disrupted by oligos 1, 2, 3 and 3a but not by oligos 2a, 2b and 3b. Oligo 4, which targets the second stem-loop (SLII) in the cIAP1 IRES, also masks NF45 binding partially. We predict that this stem-loop may be required for optimal NF45–RNA complex formation.
In examining published nucleic acid motifs that interact with NF45 and/or NF90, we noted that the structure encapsidation signal RNA of hepatitis B virus (HBVɛ) resembles stem-loop I of the cIAP1 IRES including a prominent bulge. Shin et al.33 showed that NF45/NF90 interaction with HBVɛ RNA is dependent on the bulge of its stem-loop. We performed a similar experiment by deleting the CUUA nucleotide sequence that comprises the bulge within the cIAP1 IRES using site-directed mutagenesis to create Probe 3ΔCUAA. Equivalent amounts of labeled RNA probe corresponding to the wild-type or CUUA mutant IRES were incubated with recombinant GST–NF45 or GST alone, followed by UV cross-linking and resolving the complex by SDS-PAGE. We found that the bulge in stem-loop I was not required for NF45 binding to the cIAP1 IRES (Figure 5d).
The results from our oligo competition experiment suggested that the AU-rich base of stem-loop I was important for binding to NF45. To directly confirm this hypothesis, we synthesized a new RNA probe corresponding to stem-loop I of the cIAP1 IRES (probe SLIwild type) as well as two mutant probes that could distinguish between sequence (SLIs) and sequence+structure-dependent (SLIss) interactions. We found that NF45 interacts with the wild-type SLI RNA but not SLIs or SLss mutants (Figure 5d). Together with the oligo masking data (specifically the lack of binding observed with oligo 2 that disrupts SL1 structure), we conclude that NF45 binds in a sequence and structure-specific manner to the AU stem-loop present in the cIAP1 IRES.
Discussion
Unlike cellular IRES, viral IRES exhibit conserved secondary structures that, with some exceptions, require minimal accessory factors to mediate recruitment of the ribosome.21 This is in contrast to the limited homology shared by the published set of cellular IRES secondary structures.34, 35 Indeed, the primary sequence and our empirically derived secondary structure of the cIAP1 IRES share no obvious similarity with the XIAP IRES and other published cellular IRES structures. Although cellular IRES comprise an evolutionarily divergent group, specific cellular RNA structure motifs have likely been co-opted by viruses at an early stage in evolution that allows them to compete for host ribosomes successfully. Meanwhile, the emerging need for eukaryotes to control protein synthesis in time and space may have lead to a greater dependence on ITAFs, which can modulate IRES activity of a diverse set of mRNAs in different physiological contexts. Indeed we found that cIAP1 IRES is unable to interact with the purified 40S ribosome in the absence of other protein factors.
In a search for protein factors that modulate translation of cIAP1 mRNA, we identified a cohort of RNA binding proteins, using the cIAP1 IRES as an RNA affinity probe. This complement of cIAP1 IRES binding proteins includes RNA helicase A, IGF2BP1, NF90 and NF45. The cIAP1 IRES is induced during ER stress, therefore we had initially attempted to compare protein–IRES interactions between cells treated with and without tunicamycin – a pharmacological inducer of ER stress. However, one-dimensional RNA affinity chromatography failed to reveal any differences in protein affinities and/or profiles following induction of ER stress (data not shown). This points to the possibility that the regulatory event that induces IRES activity during ER stress involves either post-translational modifications or subcellular relocalization of cIAP1 ITAFs. Nevertheless, the protein complex that interacts with the cIAP1 IRES merits further investigation as all four of these proteins have been previously identified to interact with viral IRES elements, including those encoded by HRV2, hepatitis C virus (HCV) and bovine viral diarrhea virus (BVDV) genomes.25, 36, 37 Importantly, no role for NF45 in translation of cellular mRNAs has been described. Interestingly, NF45/NF90 heterodimers were found to bind to the HRV2 IRES and inhibit translation of downstream cistrons.26 It appears that inhibition of IRES activity by this binary complex is highly dependent on cell type. Merrill et al.25 found that neuronal cells (including HEK293T) highly express both NF45 and NF90 that readily form heterodimers, and interact with the HRV2 IRES, leading to inhibition of downstream ORF expression. In contrast, cells from glial lineages were poor expressors of both NF45 and NF90 and showed impaired formation of functional inhibitory complexes. Complicating the study of NF45 or NF90 in isolation is the finding by Guan et al.24 that targeting NF45 expression by RNA interference significantly decreases the stability of NF90 and vice versa. Therefore, we first looked at the function of NF45 in a glial cell line in which expression levels of NF45 and NF90 are limited.
In dissecting the individual function of NF45 in U373 glioblastoma cells, we have generated data in support of the hypothesis that NF45 is involved in the regulation of cellular IRES activity and by extension, cIAP1 protein levels. We have shown that NF45 is able to enhance translation of endogenous cIAP1 mRNA when transiently expressed in U373 glioblastoma cells. Importantly, NF45 overexpression did not significantly affect steady-state cIAP1 mRNA levels, suggesting that increased translation rates are not the result of enhanced transcription or stability of cIAP1 mRNA. Indeed, we found that the cIAP1 mRNA pool was present in higher-order polysomes in cells expressing NF45, indicating that NF45 functions at the level of translation initiation. This report is the first to ascribe a direct role for NF45 in modulating the translation of a cellular mRNA, although circumstantial evidence of NF45 interacting with components of the canonical translational machinery (e.g., eIF2α/β/γ) exists in the literature.38
We identified NF45 interacting with the portion of the cIAP1 5′ UTR that confers IRES activity, therefore we surmised that the NF45-mediated changes in endogenous cIAP1 mRNA translation may be the result of NF45's ability to modulate cIAP1 IRES activity. To test this hypothesis we used a bicistronic expression vector previously used by our laboratory to assess cIAP1 IRES activity. Relative to cells expressing GFP, NF45-expressing cells exhibited a 240% increase in IRES activity indicating that NF45 modulates the translational efficiency of cIAP1 mRNA through its IRES. NF45 can therefore be categorized as a bona fide ITAF by virtue of its ability to modulate IRES activity.
To further delineate the specificity of NF45 ITAF activity we looked at the activity of several cellular IRES in HeLa cell lines that constitutively express shRNA targeting NF45 (d5) relative to cells that express a nontargeting shRNA (c). In these cells that have significantly reduced levels of NF45 expression, the activity of cIAP1 was significantly reduced, whereas other IRES (APAF-1, BclxL, DAP5, VCIP) exhibited no change or modest increase in activity (Figure 4a). These data point to the specificity of NF45 in modulating cIAP1 translation through its IRES.
We and others have shown that the cIAP1 IRES is induced as part of the UPR following ER stress. Therefore, the observed decrease in cIAP1 IRES activity in d5 cells lead us to the hypothesis that these cells should be refractory to cIAP1 induction following ER stress. Indeed, after the treatment of cells with thapsigargin we observed an induction of cIAP1 in wild-type HeLa cells but not in cells stably expressing NF45 shRNA (Figure 4b). Interestingly, thapsigargin also failed to induce cIAP1 in our U373 glioblastoma cells (expressing low levels of NF45) despite robust induction of the ER stress marker BiP (data not shown). Furthermore, we were unable to rescue the phenotype of increased cIAP1 translation during the UPR with the introduction of ectopic NF90 in d5 cells (Figure 4c). In contrast, reintroduction of RNAi-resistant NF45 into these cells induced endogenous levels of cIAP1 as expected (Figure 4c). These data suggest that the interaction of NF45 with its in vivo binding partner NF90 is not required for NF45 ITAF activity, although the data do not preclude the possibility that NF90 could act as a sink for NF45, thus reducing the effective ITAF activity of NF45 in the cell. The observation that overexpression of NF45 in the absence of ER stress can increase cIAP1 translation (in both d5 and U373 cells) would argue for the ‘sink’ hypothesis, whereby NF45 ITAF activity is masked within a stable NF45/NF90 heterodimer until a trigger (e.g., ER stress) releases it.
We next looked at physical interactions of NF45 with the cIAP1 IRES in vitro to shed light on the mechanism by which NF45 may be affecting cIAP1 IRES activity. Although there is no evidence in the current literature that NF45 interacts with RNA directly, the protein does contain two potential RNA binding domains, namely a DNA zinc-finger motif and an N-terminal RGG box motif. Importantly, we were able to resolve an NF45–IRES complex following photocross-linking of recombinant full-length NF45 and in vitro transcribed and labeled RNA corresponding to the cIAP1 IRES sequence. This shows that NF45 alone is sufficient to bind to the cIAP1 IRES in a sequence-dependent manner.
To determine the minimal NF45 binding site on the cIAP1 IRES, we used DNA oligonucleotide competitors to block potential binding sites within the IRES. From this data, we concluded that NF45 binding likely requires both sequence and structural motifs present in the base of the first stem-loop of the cIAP1 IRES. We confirmed this hypothesis by mutational analysis of stem-loop I. By mutating the AU-rich stem to a GC-rich stem, we were able to retain its structure but abrogate NF45 binding, suggesting that either the unstructured AAAA/UUUU stretch or the structured AU stem-loop was necessary (Figure 5d, mutant SLIs). The efficient masking of binding with oligo 2 (that prevents formation of the AU stem-loop but preserves access to the single-stranded AAAA/UUUU sequence) from our oligo competition experiment allows us to conclude that primary sequence (i.e., AAAA/UUUU) alone is not sufficient for binding of NF45, rather it is the base of the stem-loop formed by AU base pairs that are required. The lack of binding that we observed with a mutant that disrupts both sequence and structure in this region (SLIss) supports this conclusion. Further, stem-loop II of the IRES contains asymmetrical AU base pairs that may explain the partial binding with NF45 that we observed when using probes 2 and 3 that have stem-loop II in common (Figure 5b) and when masking with oligo 4 (Figure 5c ). Intriguingly, the AU content of the UTRs that exhibited no change or increased IRES activity in NF45-deficient cells (Figure 4a) was significantly lower than that for cIAP1 (cIAP1, 70%; APAF1, 32%; BclxL, 46%; DAP5, 43%; VCIP, 34%). These observations suggest that the AU stem-loop motif is necessary for NF45 ITAF activity in the context of cIAP1.
Our interpretations agree with data from other groups who have investigated NF45 interactions with RNA. For example, using mutational analysis, Isken et al.36 determined that the interaction of the NF45/NF90 proteins with HCV and BVDV 5′ and 3′ UTRs was dependent on both sequence and stem-loop structures. In another study NF45 and NF90 were found to interact with the HBVɛ RNA.33 The authors showed that NF45/NF90 interaction with HBVɛ RNA is dependent on the bulge of its stem-loop redundant.33 We noticed that the HBVɛ structure resembles the first stem-loop of the cIAP1 IRES including a prominent bulge. On the basis of this, we hypothesized that this bulge would also be important for the NF45–cIAP1 IRES interaction. Surprisingly, we found that the bulge mutant (Figure 5d, mutant 3ΔCUUA) showed no impairment in binding to NF45. This data imply that the NF45/NF90 heterodimer and NF45 alone have different sequence requirements for binding to RNA.
The specific domain(s) that is responsible for interaction of NF45 with the cIAP1 IRES remains unknown. A likely candidate is the conserved RGG box motif at the N terminus of the protein. RGG box motifs can serve as RNA binding domains and are present in many RNA binding proteins; therefore the potential importance of the RGG box motif in the context of NF45 and cIAP1 IRES interactions merits further investigation. The RGG box motifs in nucleolin and hnRNPU have been shown to disrupt RNA secondary structure in a nonspecific manner.39, 40 For other proteins such as FMRP, the RGG box imparts a more specific RNA binding activity. Specifically, the RGG box within FMRP confers binding to planar RNA conformations referred to as G-quartet structures within mRNAs.41, 42 The idea that the cIAP1 IRES can adopt an inter- or intramolecular G-quartet structure that can have a role in translational regulation is an intriguing possibility. Whether the NF45 RGG box could participate in a manner analogous to FMRP is an open question that is currently under investigation.
The mechanism that the cIAP1–ITAF complex may use to modulate translation remains to be elucidated; however we have shown that NF45, a component of this complex, is able to interact directly with the cIAP1 IRES and positively affect its activity, resulting in increased translation of cIAP1 mRNA. Thus, NF45 is a novel RNA binding protein and ITAF that functions to upregulate cIAP1 translation during ER stress.
Materials and Methods
Cell culture, expression constructs and transfection
Human embryonic kidney (HEK293T) and human glioblastoma (U373 MG) cells were maintained in standard conditions in serum- and antibiotic-supplemented Dulbecco's modified Eagle's medium (DMEM). c and d5 HeLa cell lines were previously characterized.24 HeLa cells expressing shRNA were maintained under selective pressure with 400 μg/ml of G418 (Invitrogen, Burlington, ON, Canada). For transient knockdown of NF45, we transfected 293T cells with two rounds of 50 nM d5 siRNA and Lipofectamine 2000 (Invitrogen) over a period of 96 h. The full-length NF45 ORF from an NF45 expression plasmid (pEF.NF45.HA; Kao et al.20) was re-cloned into a pcDNA3 backbone as an N-terminal FLAG-tagged vector (pcDNA3.F.NF45) and verified by sequencing. Transient transfections of U373 MG cells were performed using Amaxa nucleofector technology (Lonza, Germany; solution T, program T-020). Transfection efficiency was verified by light microscopy, western blotting and RT-PCR. For d5 rescue experiments, 2 μg O.NF90c24 or F.NF45r (an RNAi-resistant version of F.NF45 in which a silent mutation was introduced by SDM) DNA was transfected using Lipofectamine 2000 (Invitrogen). Unless otherwise stated, all assays were performed 24 h after transfection.
RNA secondary structure determination
The cIAP1 IRES secondary structure was determined using enzymatic probing with RNases as described previously.34 RNase cut sites were used as constraints in either mfold22 or RNAStructure43 to predict secondary structure models. The RNA secondary structure graphic was generated using jViz.Rna (Available at: http://jviz.cs.sfu.ca).
In vitro transcription
DNA templates for the synthesis of the cIAP1 RNA probes were generated from a cIAP1 5′ UTR-containing plasmid (probes 1–3) or synthesized oligonucleotides (for SLI probes) by PCR. The 5′ primers incorporated the T7 promoter sequence. For UV cross-linking experiments, we synthesized labeled RNA probes by in vitro transcription with T7 polymerase (MAXIscript kit; Applied Biosystems, Streetsville, ON, Canada) in the presence of [α-32P]UTP (PerkinElmer, Woodbridge, ON, Canada) followed by gel purification of full-length transcripts. For RNA affinity chromatography, unlabeled RNA was synthesized using T7 polymerase (MEGAshortscript; Applied Biosystems).
RNA affinity chromatography
For preparation of cytoplasmic lysates, confluent HEK293T cells were resuspended in lysis buffer (10 mM Tris-HCl (pH 7.4), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.1 mM PMSF, 10 μg/ml leupeptin) and dounce-homogenized. Lysates were centrifuged at 10 000 × g for 10 min at 4°C, and the supernatant was retained. Protein concentration was determined using a modified Lowry Assay (DC Protein Assay; Bio-Rad, Mississauga, ON, Canada). In vitro synthesized cIAP1 RNA was 5′ end-labeled with biotin (5′ End Tag; Vector Laboratories, Burlington, ON, Canada), and conjugated to avidin-coated agarose beads (Sigma-Aldrich, Oakville, ON, Canada) for 2 h at 4°C in the presence of RNA binding buffer (10 mM Tris-HCl (pH 7.4), 150 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 0.05% NP-40, 10 μg/ml leupeptin and 1 U/ml RNase inhibitor). The beads were then washed twice with RNA binding buffer and incubated with 200 μg of precleared HEK293T S10 extract and 30 μg of wheat germ tRNA (Sigma-Aldrich) for 30 min at room temperature followed by an additional 2 h at 4°C. The beads were then washed five times with RNA binding buffer and bound proteins were released by boiling in Laemmli buffer. Captured proteins were resolved on 10% SDS-PAGE and stained overnight with SYPRO Ruby (Invitrogen). Specific bands were excised and analyzed by MALDI-TOF mass spectrometry and peptide mass fingerprinting (Queen's University Protein Function Discovery Facility, Kingston, Canada).
Western blot analysis
Cells were lysed in RIPA buffer for 30 min at 4°C, followed by centrifugation at 10 000 × g to remove debris. Equal amounts of protein were resolved by 10% SDS-PAGE, transferred to PVDF membranes using a semidry transfer protocol and probed with antibodies to RNA Helicase A (Center for Biomedical Inventions, University of Texas Southwestern at Dallas), NF90 (anti-DRBP76; BD Laboratories or anti-NF9044), IGF2BP1 (anti-IMP1; a gift from Dr. F Nielsen), NF45,44 HuR (clone 3A2; Santa Cruz Biotechnology), TIA-1/TIAR (clone 3E6; a gift from Dr. P Anderson), hnRNPA1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), β-actin (Sigma-Aldrich) and GAPDH (BD Laboratories, Mississauga, ON, Canada). Membranes were then incubated with species-specific horseradish-peroxidase-conjugated secondary antibody (GE Biosciences, Baie d'Urfe, QC, Canada) followed by detection with ECL+ substrate (GE Biosciences). Alternatively, membranes were incubated with Alexa 680-conjugated (Invitrogen) or IR 800-conjugated (LI-COR Biotechnology, Lincoln, NE, USA) secondary antibody followed by detection using the Licor Odyssey Infrared scanner.
Metabolic labeling/immunoprecipitation
A total of 1.5 × 106 U373 MG cells were nucleofected with 10 μg of GFP or NF45 DNA plasmid and seeded in 10 cm plates 24 h before assay. Cells were incubated with DMEM lacking methionine and cysteine and supplemented with 10% FCS (Invitrogen) for 15 min at 37°C. Cells were pulse-labeled with 0.1 mCi/ml 35S-methionine and cysteine mix (EasyTag EXPRE35S35S Protein Labelling Mix; PerkinElmer) for 25 min at 37°C. Cells were washed, harvested in cold PBS and boiled in 50 μl of denaturing lysis buffer (50 mM Tris (pH 7.4), 5 mM EDTA, 1% SDS, 10 mM DTT, 1 mM PMSF, 2 μg/ml leupeptin, 15 U/ml DNaseI) for 5 min. Cold nondenaturing buffer (450 μl; 50 mM Tris (pH 7.4), 5 mM EDTA, 300 mM NaCl, 1% Triton X-100, 10 mM iodoacetamide, 1 mM PMSF, 2 μg/ml leupeptin) was then added and the lysate was passed through a 25-gauge syringe needle 10 times. Following centrifugation to remove debris, we precleared the lysate before immunoprecipitation with Pansorbin cells for 2 h at 4°C (EMD Chemicals, Gibbstown, NJ, USA). Co-immunoprecipitation of cIAP1 and β-actin from the precleared lysates was performed at 4°C for 16 h using Protein G/Protein A-Agarose beads (EMD Chemicals) coated with antibodies specific for β-actin (Sigma-Aldrich) and cIAP145 at a titer of 1 : 500 and 1 : 150, respectively. The beads were then washed extensively with cold wash buffer (50 mM Tris (pH 7.4), 300 mM NaCl, 0.1% Triton X-100), resuspended in Laemmli buffer and boiled to elute bound proteins. Immunoprecipitated proteins or total proteins were then resolved on 10% SDS-PAGE. The gel was incubated with Amplify fluorogenic reagent (GE Biosciences) for 30 min before drying and exposure to film. Densitometric analysis was performed using Licor Odyssey software (LI-COR Biotechnology).
Quantitative RT-PCR
To measure relative mRNA expression, total RNA was isolated from U373MG cells 24 h after transient expression of GFP or NF45 (Absolutely RNA miniprep; Agilent Technologies, La Jolla, CA, USA) and cDNA generated (First-Strand cDNA synthesis kit; GE Biosciences). Quantitative PCR was performed on an ABI Prism 7000 real-time thermocycler using 1 μg total RNA together with SYBRGreen (Qiagen, Mississauga, ON, Canada) and gene-specific primers (cIAP1: TCTGGAGATGATCCATGGGTAGA, TGGCCTTTCATTCGTATCAAGA; β-actin: CTGGAACGGTGAAGGTGACA, AAGGGACTTCCTGTAACAATGCA; NF45: GACACAATGTGGCTGACCTG, GAAGATTGGGTGGCACTGTT). Relative expression levels were determined using the standard curve method. Controls lacking RT showed no significant genomic DNA amplification (>10 cycle difference).
Polysome profiling
A total of 5 × 106 U373 MG cells were nucleofected with 10 μg of GFP or NF45 plasmid DNA 72 h before polysome profiling. Cells were incubated with 0.1 mg/ml cycloheximide for 3 min, washed with cold PBS+cycloheximide and lysed in cold polysome lysis buffer (15 mM Tris-HCl (pH 7.4), 15 mM MgCl2, 300 mM NaCl, 1% (v/v) Triton X-100, 0.1 mg/ml cycloheximide, 100 U/ml RNasin). Equal OD254 units were loaded onto 10–50% linear sucrose gradients and centrifuged at 39 000 r.p.m. for 90 min at 4°C. Gradients were fractionated from the top (Densi-Flow; Labconco, Kansas City, MO, USA) and RNA/protein was monitored at 254 nm using a HPLC system (Akta Explorer; GE Biosciences). Fractions (1 ml) were collected and flash-frozen in liquid nitrogen. RNA was isolated from individual fractions by proteinase K digestion followed by phenol/chloroform extraction. Equal volumes of RNA from each fraction were used to generate cDNA using oligo-dT primers and a reverse transcription kit (First-Strand cDNA synthesis kit; GE Biosciences). PCR primers specific for cIAP1 or β-actin (cf. last section for specific sequences) were used to amplify messages using a limited-cycle PCR. Amplified cDNA was resolved on agarose gel, and visualized with ethidium bromide. Genomic DNA contamination in controls lacking RT was undetectable.
IRES activity assay
A total of 1 × 106 U373 MG cells were nucleofected with 6 μg βGAL/cIAP1/CAT bicistronic vector containing the full-length cIAP1 5′ UTR (nucleotides −1222 to −1) and 2 μg GFP or NF45 expression vector. For monocistronic experiments, CAT expression constructs with the 1.2 kb 5′ UTR or a mutant containing nucleotides −80 to −1 of the 5′ UTR were used together with a β-GAL expression construct. For c and d5 HeLa cell experiments, we seeded 6 × 105 cells 24 h before Lipofectamine 2000 (Invitrogen) transfection with 2 μg bicistronic vectors harboring cIAP1, APAF-1, BclxL, DAP5 and VCIP IRES. Protein lysates were harvested after 24 h using CAT lysis buffer (Roche Diagnostics, Laval, QC, Canada). β-GAL activity was assessed using an ONPG colorimetric assay. CAT expression was quantified by ELISA according to the manufacturer's protocol (Roche Diagnostics). Spurious splicing and potential cryptic promoter activity that could arise with the pBGAL/cIAP1/CAT bicistronic vector has been previously addressed.27
Recombinant NF45 expression and purification
The full-length ORF of human NF45 was amplified from pEF.NF45.HA by PCR and cloned into a bacterial GST expression plasmid (pGEX-KG). Expression of GST-NF45 (72 kDa) was induced with 100 nM IPTG for 4 h at 37°C. Cells were lysed on ice with lysozyme, 1.5% Sarkosyl, DNase I and protease inhibitors. Triton X-100 (4%) was added to solubilize proteins before clarification by centrifugation. Lysates were incubated with Glutathione Sepharose 4B beads (GE Biosciences) for 16 h and GST moieties were eluted with reduced glutathione (20 mM glutathione, 30 mM Na2HPO4, 0.1% (w/v) CHAPS, pH 9.5) for 2 h at 4°C. Purity was assessed by SDS-PAGE and Coomassie staining.
RNA UV cross-linking
[α-32P]UTP-labeled, in vitro transcribed cIAP1 RNA (40 000 c.p.m.) was incubated with 300–500 ng of GST-NF45 for 25 min at room temperature in an RNA binding buffer (10 mM Tris-HCl (pH 7.4), 1.5 mM MgCl2, 150 mM KCl, 0.5 mM DTT, 0.1 mM PMSF, 10 mg/ml leupeptin). Samples were UV-irradiated (250 mJ/cm2) on ice using a Stratagene Stratalinker, followed by treatment with 2 U of RNase A/T1 and 100 μg of heparin. Complexes were resolved by SDS-PAGE and visualized by autoradiography.
Conflict of interest
The authors declare no conflict of interest.
Abbreviations
- cIAP1:
-
cellular inhibitor of apoptosis protein 1
- ER:
-
endoplasmic reticulum
- IRES:
-
inducible internal ribosome entry site
- NF45:
-
nuclear factor 45
- UTR:
-
5′ untranslated region
References
Qin X, Sarnow P . Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 2004; 279: 13721–13728.
Gerlitz G, Jagus R, Elroy-Stein O . Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur J Biochem 2002; 269: 2810–2819.
Bushell M, Sarnow P . Hijacking the translation apparatus by RNA viruses. J Cell Biol 2002; 158: 395–399.
Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcik M . The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res 2008; 36: 168–178.
Yoshimura FK, Luo X, Zhao X, Gerard HC, Hudson AP . Up-regulation of a cellular protein at the translational level by a retrovirus. Proc Natl Acad Sci USA 2008; 105: 5543–5548.
Koritzinsky M, Magagnin M, Van Den Beucken T, Seigneuric R, Savelkouls K, Dostie J et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J 2006; 25: 1114–1125.
Yang DQ, Halaby MJ, Zhang Y . The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 2006; 25: 4613–4619.
Pestova TV, Lorsch JR, Hellen CUT . Translational control in biology and medicine. In: Mathews M, Sonenberg N, Hershey JWB (eds). Cold Spring Harbor Monograph Series. 3rd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2007. pp 87–128.
Holcik M, Sonenberg N . Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6: 318–327.
Bushell M, Stoneley M, Kong Y, Hamilton T, Spriggs K, Dobbyn H et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell 2006; 23: 401–412.
Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M . Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell 2007; 18: 1302–1311.
Holcik M, Korneluk RG . Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol 2000; 20: 4648–4657.
Holcik M, Gordon BW, Korneluk RG . The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol Cell Biol 2003; 23: 280–288.
Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP et al. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev 2005; 19: 1556–1571.
Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 2003; 113: 519–531.
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693.
Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007; 131: 669–681.
Warnakulasuriyarachchi D . Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem 2004; 279: 17148–17157.
Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE . Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA 2004; 10: 469–481.
Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem 1994; 269: 20691–20699.
Baird SD, Turcotte M, Korneluk RG, Holcik M . Searching for IRES. RNA 2006; 12: 1755–1785.
Zuker M . Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31: 3406–3415.
Jan E, Sarnow P . Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 2002; 324: 889–902.
Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol 2008; 28: 4629–4641.
Merrill MK, Dobrikova EY, Gromeier M . Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol 2006; 80: 3147–3156.
Merrill M . The double-stranded RNA binding protein 76: NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol 2006; 80: 6936–6942.
Holcik M . Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the gal/CAT bicistronic reporter system. RNA 2005; 11: 1605–1609.
Ungureanu NH, Cloutier M, Lewis SM, de Silva N, Blais JD, Bell JC et al. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J Biol Chem 2006; 281: 15155–15163.
Yoon A . Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006; 312: 902–906.
Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 2008; 30: 447–459.
Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol 2006; 26: 9517–9532.
Ranpura SA, Deshmukh U, Reddi PP . NF45 and NF90 in murine seminiferous epithelium: potential role in SP-10 gene transcription. J Androl 2008; 29: 186–197.
Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM . Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch Virol 2002; 147: 471–491.
Baird SD, Lewis SM, Turcotte M, Holcik M . A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res 2007; 35: 4664–4677.
Xia X, Holcik M . Strong eukaryotic IRESs have weak secondary structure. PLoS ONE 2009; 4: e4136.
Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A et al. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 2007; 13: 1675–1692.
Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F et al. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J 2003; 22: 5655–5665.
Ting NS, Kao PN, Chan DW, Lintott LG, Lees-Miller SP . DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J Biol Chem 1998; 273: 2136–2145.
Kiledjian M, Dreyfuss G . Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J 1992; 11: 2655–2664.
Ghisolfi L, Joseph G, Amalric F, Erard M . The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem 1992; 267: 2955–2959.
Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB . Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001; 107: 489–499.
Ramos A, Hollingworth D, Pastore A . G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA 2003; 9: 1198–1207.
Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH . Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA 2004; 101: 7287–7292.
Parrott AM, Walsh MR, Reichman TW, Mathews MB . RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J Mol Biol 2005; 348: 281–293.
Zhao TT, Graber TE, Jordan LE, Cloutier M, Lewis SM, Goulet I et al. hnRNP A1 regulates UV-induced NF-kappaB signalling through destabilization of cIAP1 mRNA. Cell Death Differ 2009; 16: 244–252.
Acknowledgements
We thank Dr. E Jan for the help with the ribosome electromobility shift assay. This work was supported by an operating grant from the Canadian Institutes for Health Research (FRN 74740) to MH and from the National Institutes of Health (NIH AI 034552) to MBM. TEG was supported by the Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Award. MH is the CHEO Volunteer Association Endowed Scholar.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by V Dixit
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Graber, T., Baird, S., Kao, P. et al. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ 17, 719–729 (2010). https://doi.org/10.1038/cdd.2009.164
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.164
Keywords
This article is cited by
-
Aberrant accumulation of NIK promotes tumor growth by dysregulating translation and post-translational modifications in breast cancer
Cancer Cell International (2023)
-
Quercetin induces pannexin 1 expression via an alternative transcript with a translationally active 5′ leader in rhabdomyosarcoma
Oncogenesis (2022)
-
Identification of cryptic putative IRESs within the ORF encoding the nonstructural proteins of the human rhinovirus 16 genome
Archives of Virology (2021)
-
Eukaryotic initiation factor 5B (eIF5B) provides a critical cell survival switch to glioblastoma cells via regulation of apoptosis
Cell Death & Disease (2019)
-
Identification of host cellular proteins that interact with the M protein of a highly pathogenic porcine reproductive and respiratory syndrome virus vaccine strain
Virology Journal (2017)