Abstract
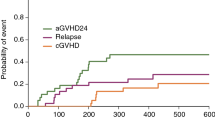
The identification of important amino acid substitutions associated with low survival in hematopoietic cell transplantation (HCT) is hampered by the large number of observed substitutions compared with the small number of patients available for analysis. Random forest analysis is designed to address these limitations. We studied 2107 HCT recipients with good or intermediate risk hematological malignancies to identify HLA class I amino acid substitutions associated with reduced survival at day 100 post transplant. Random forest analysis and traditional univariate and multivariate analyses were used. Random forest analysis identified amino acid substitutions in 33 positions that were associated with reduced 100 day survival, including HLA-A 9, 43, 62, 63, 76, 77, 95, 97, 114, 116, 152, 156, 166 and 167; HLA-B 97, 109, 116 and 156; and HLA-C 6, 9, 11, 14, 21, 66, 77, 80, 95, 97, 99, 116, 156, 163 and 173. In all 13 had been previously reported by other investigators using classical biostatistical approaches. Using the same data set, traditional multivariate logistic regression identified only five amino acid substitutions associated with lower day 100 survival. Random forest analysis is a novel statistical methodology for analysis of HLA mismatching and outcome studies, capable of identifying important amino acid substitutions missed by other methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood 2004; 104: 1923–1930.
Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007; 110: 4576–4583.
Shaw BE . The clinical implications of HLA mismatches in unrelated donor haematopoietic cell transplantation. Int J Immunogenet 2008; 35: 367–374.
Hauzenberger D, Schaffer M, Ringdén O, Hassan Z, Omazic B, Mattsson J et al. Outcome of haematopoietic stem cell transplantation in patients transplanted with matched unrelated donors vs allele-mismatched donors: a single centre study. Tissue Antigens 2008; 72: 549–558.
Petersdorf EW . Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol 2008; 20: 588–593.
Whitelegg A, Barber LD . The structural basis of T-cell allorecognition. Tissue Antigens 2004; 63: 101–108.
Archbold JK, Ely LK, Kjer-Nielsen L, Burrows SR, Rossjohn J, McCluskey J et al. T cell allorecognition and MHC restriction-A case of Jekyll and Hyde. Mol Immunol 2008; 45: 583–598.
Keever CA, Leong N, Cunningham I . HLA-B44-directed cytotoxic T cells associated with acute graft-versus-host disease following unrelated bone marrow transplantation. Bone Marrow Transplant 1994; 14: 137–145.
Fleischhauer K, Kernan NA, O’Reilly RJ, Dupont B, Yang SY . Bone marrow-allograft rejection by T lymphocytes recognizing a single amino acid difference in HLA-B44. N Engl J Med 1990; 323: 1818–1822.
Ferrara GB, Bacigalupo A, Lamparelli T, Lanino E, Delfino L, Morabito A et al. Bone marrow transplantation from unrelated donors: the impact of mismatches with substitutions at position 116 of the human leukocyte antigen class I heavy chain. Blood 2001; 98: 3150–3155.
Kawase T, Morishima Y, Matsuo K, Kashiwase K, Inoko H, Saji H et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood 2007; 110: 2235–2241.
Heemskerk MB, Roelen DL, Dankers MK, van Rood JJ, Claas FH, Doxiadis II et al. Allogeneic MHC class I molecules with numerous sequence differences do not elicit CTL response. Human Immunol 2005; 66: 969–976.
Heemskerk MB, Cornelissen JJ, Roelen DL, van Rood JJ, Claas FH, Doxiadis II et al. Highly diverged MHC class I mismatches are acceptable for haematopoietic stem cell transplantation. Bone Marrow Transplant 2007; 40: 193–200.
Breiman L . Random Forests. Machine Learning J 2001; 45: 5–32.
Breiman L . Statistical modeling: the two cultures. Statist Sci 2001; 16: 199–231.
Lunetta KL, Hayward LB, Segal J, Van Eerdewegh P . Screening large-scale association study data: exploiting interactions using random forests. BMC Genet 2004; 5: 32.
Díaz-Uriarte R, de Andrés SA . Gene selection and classification of microarray data using random forest. BMC Bioinformatics 2006; 7: 3.
Dudoit S, Fridlyand J, Speed TP . Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc 2002; 97: 77–87.
Lee JW, Lee JB, Park M, Song SH . An extensive evaluation of recent classification tools applied to microarray data. Comput Stat Data Anal 2005; 48: 869–885.
Dew A, Collins D, Artz A, Rich E, Stock W, Swanson K et al. Paucity of HLA-identical unrelated donors for African-Americans with hematologic malignancies: the need for new donor options. Biol Blood Marrow Transplant 2008; 14: 938–941.
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC . Structure of the human class I histocompatibility antigen, HLA-A2. Nature 1987; 329: 506–512.
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC . The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 1987; 329: 512–518.
Bjorkman PJ, Strominger JL, Wiley DC . Crystallization and X-ray diffraction studies on the histocompatibility antigens HLA-A2 and HLA-A28 from human cell membranes. J Mol Biol 1985; 186: 205–210.
Burrows SR, Khanna R, Burrows JM, Moss DJ . An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTL) cross-reactive with a single Epstein-Barr virus CTL epitope. implications for graft-versus-host disease. J Exp Med 1994; 179: 1155–1161.
Kawase T, Matsuo K, Kashiwase K, Inoko H, Saji H, Ogawa S et al. HLA mismatch combinations associated with decreased risk of relapse: Implications for molecular mechanism. Blood 2009; 113: 2851–2858.
Acknowledgements
We thank Theodore Karrison, PhD, for statistical support. This study was supported by the University of Chicago Cancer Research Center, Chicago, Illinois (Fund-6-33573 (SRM)). The Center for International Blood and Marrow Transplant Research is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases; a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (DHHS); two Grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from AABB; Aetna; American Society for Blood and Marrow Transplantation; Amgen Inc.; Anonymous donation to the Medical College of Wisconsin; Astellas Pharma US Inc.; Baxter International Inc.; Bayer HealthCare Pharmaceuticals; Be the Match Foundation; Biogen IDEC; BioMarin Pharmaceutical Inc.; Biovitrum AB; BloodCenter of Wisconsin; Blue Cross and Blue Shield Association; Bone Marrow Foundation; Canadian Blood and Marrow Transplant Group; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Centers for Disease Control and Prevention; Children's Leukemia Research Association; ClinImmune Labs; CTI Clinical Trial and Consulting Services; Cubist Pharmaceuticals; Cylex Inc.; CytoTherm; DOR BioPharma Inc.; Dynal Biotech, an Invitrogen Company; Eisai Inc.; Enzon Pharmaceuticals Inc.; European Group for Blood and Marrow Transplantation; Gamida Cell Ltd.; GE Healthcare; Genentech Inc.; Genzyme Corporation; Histogenetics Inc.; HKS Medical Information Systems; Hospira Inc.; Infectious Diseases Society of America; Kiadis Pharma; Kirin Brewery Co. Ltd.; The Leukemia and Lymphoma Society; Merck and Company; The Medical College of Wisconsin; MGI Pharma Inc.; Michigan Community Blood Centers; Millennium Pharmaceuticals Inc.; Miller Pharmacal Group; Milliman USA Inc.; Miltenyi Biotec Inc.; National Marrow Donor Program; Nature Publishing Group; New York Blood Center; Novartis Oncology; Oncology Nursing Society; Osiris Therapeutics Inc.; Otsuka America Pharmaceutical Inc.; Pall Life Sciences; PDL BioPharma Inc; Pfizer Inc; Pharmion Corporation; Saladax Biomedical Inc.; Schering Corporation; Society for Healthcare Epidemiology of America; StemCyte Inc.; StemSoft Software Inc.; Sysmex America Inc.; Teva Pharmaceutical Industries; THERAKOS Inc.; Thermogenesis Corporation; Vidacare Corporation; Vion Pharmaceuticals Inc.; ViraCor Laboratories; ViroPharma Inc.; and Wellpoint Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the US Government.
Author contributions: SRM conceptualized the study, interpreted the results and wrote the manuscript; SRM and SL designed the study; SL performed the univariate, multivariate, and random forest analyses; MM prepared amino acid database for analysis; MH prepared data for statistical analysis; SS and SJL contributed ideas and made significant contributions to the writing of the manuscript; JK performed multivariate analysis; TAB prepared the figure; KVB provided overall advice and guidance. All authors reviewed the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Marino, S., Lin, S., Maiers, M. et al. Identification by random forest method of HLA class I amino acid substitutions associated with lower survival at day 100 in unrelated donor hematopoietic cell transplantation. Bone Marrow Transplant 47, 217–226 (2012). https://doi.org/10.1038/bmt.2011.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bmt.2011.56
Keywords
This article is cited by
-
Multistate recursively imputed survival trees for time-to-event data analysis: an application to AIDS and mortality post-HIV infection data
BMC Medical Research Methodology (2018)
-
Identification of high-risk amino-acid substitutions in hematopoietic cell transplantation: a challenging task
Bone Marrow Transplantation (2016)