Abstract
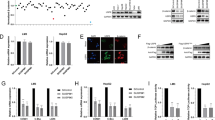
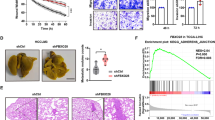
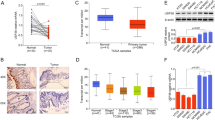
In the current study, we have shown that USP51 promotes colorectal cancer stemness and chemoresistance, and high expression of USP51 predicts survival disadvantage in colorectal cancer patients. Mechanically, USP51 directly binds to Elongin C (ELOC) and forms a larger functional complex with VHL E3 ligase (USP51/VHL/CUL2/ELOB/ELOC/RBX1) to regulate the ubiquitin-dependent proteasomal degradation of HIF1A. USP51 efficiently deubiquitinates HIF1A and activates hypoxia-induced gene transcription. Conversely, the activation of HIF1A under hypoxia transcriptionally upregulates the expression of USP51. Thus, USP51 and HIF1A form a positive feedback loop. Further, we found that the SUMOylation of ELOC at K32 inhibits its binding to USP51. SUMO-specific protease 1 (SENP1) mediates the deSUMOylation of ELOC, promoting the binding of USP51 to ELOC and facilitating the deubiquitination and stabilization of HIF1A by USP51. Importantly, USP51 plays a crucial role in promoting the HIF1A and SENP1-dependent proliferation, migration, stemness, and chemoresistance under hypoxia in colorectal cancer. Together, our data revealed that USP51 is an oncogene stabilizing the pro-survival protein HIF1A, offering a potential therapeutic target for colorectal cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the corresponding author.
References
Lin JS, Perdue LA, Henrikson NB, Bean SI, Blasi PR. Screening for colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA. 2021;325:1978–98.
Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, et al. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 2016;30:946–59.
Zhou Z, Zhang P, Hu X, Kim J, Yao F, Xiao Z, et al. USP51 promotes deubiquitination and stabilization of ZEB1. Am J Cancer Res. 2017;7:2020–31.
Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–44.
Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–75.
Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–66.
Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–97.
El-Asmi F, McManus FP, Thibault P, Chelbi-Alix MK. Interferon, restriction factors and SUMO pathways. Cytokine Growth Factor Rev. 2020;55:37–47.
Yeh ET. SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem. 2009;284:8223–7.
Sun XX, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, et al. SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2018;115:10983–8.
Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–27.
Xu Z, Guo C, Ye Q, Shi Y, Sun Y, Zhang J, et al. Endothelial deletion of SHP2 suppresses tumor angiogenesis and promotes vascular normalization. Nat Commun. 2021;12:6310.
Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432.
Jiang M, Chiu SY, Hsu W. SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ. 2011;18:1005–15.
Beauclair G, Bridier-Nahmias A, Zagury JF, Saïb A, Zamborlini A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics. 2015;31:3483–91.
Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–d73.
Moon EJ, Mello SS, Li CG, Chi JT, Thakkar K, Kirkland JG, et al. The HIF target MAFF promotes tumor invasion and metastasis through IL11 and STAT3 signaling. Nat Commun. 2021;12:4308.
Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut. 2017;66:2149–59.
Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–64.
Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149:1884–95.e4.
Chen MC, Nhan DC, Hsu CH, Wang TF, Li CC, Ho TJ, et al. SENP1 participates in Irinotecan resistance in human colon cancer cells. J Cell Biochem. 2021;122:1277–94.
Shangguan X, He J, Ma Z, Zhang W, Ji Y, Shen K, et al. SUMOylation controls the binding of hexokinase 2 to mitochondria and protects against prostate cancer tumorigenesis. Nat Commun. 2021;12:1812.
Wu YC, Ling TY, Lu SH, Kuo HC, Ho HN, Yeh SD, et al. Chemotherapeutic sensitivity of testicular germ cell tumors under hypoxic conditions is negatively regulated by SENP1-controlled sumoylation of OCT4. Cancer Res. 2012;72:4963–73.
Hassin O, Nataraj NB, Shreberk-Shaked M, Aylon Y, Yaeger R, Fontemaggi G, et al. Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nat Commun. 2022;13:2800.
Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70.
Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–8.
Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–34.
Madan E, Parker TM, Pelham CJ, Palma AM, Peixoto ML, Nagane M, et al. HIF-transcribed p53 chaperones HIF-1α. Nucleic Acids Res. 2019;47:10212–34.
Tang YA, Chen YF, Bao Y, Mahara S, Yatim S, Oguz G, et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proc Natl Acad Sci USA. 2018;115:E5990–E5999.
Mu M, Yu Q, Zhang Q, Guo J, Wang X, Sun X, et al. A pan-cancer analysis of molecular characteristics and oncogenic role of gasdermins. Cancer Cell Int. 2022;22:80.
Miyamoto T, Hosoba K, Itabashi T, Iwane AH, Akutsu SN, Ochiai H, et al. Insufficiency of ciliary cholesterol in hereditary Zellweger syndrome. Embo j. 2020;39:e103499.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82002547).
Author information
Authors and Affiliations
Contributions
MM and QZ: Conceptualization, methodology, validation, original draft writing, review & editing. JL: Conducted animal experiments. YZ, XL, and ZC: Analyzed the data. XS: Conceptualization, supervision, project administration. JY: Supervision, project administration, investigation, methodology, resourcing, review & editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All related experiments in our study were carried out in accordance with the Helsinki Declaration, and approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
Informed consent
Informed consent was acquired from all participants involving human tissues.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mu, M., Zhang, Q., Li, J. et al. USP51 facilitates colorectal cancer stemness and chemoresistance by forming a positive feed-forward loop with HIF1A. Cell Death Differ 30, 2393–2407 (2023). https://doi.org/10.1038/s41418-023-01228-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01228-8