Abstract
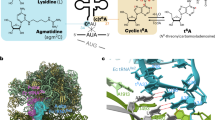
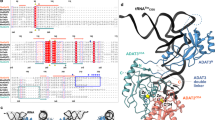
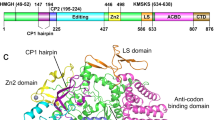
Leucyl-tRNA synthetase (LeuRS) specifically recognizes the characteristic long variable arm and the discriminator base, A73, of tRNALeu in archaea and eukarya. The LeuRS 'editing domain' hydrolyzes misformed noncognate aminoacyl-tRNA. Here we report the crystal structure of the archaeal Pyrococcus horikoshii LeuRS–tRNALeu complex. The protruding C-terminal domain of LeuRS specifically recognizes the bases at the tip of the long variable arm. The editing domain swings from its tRNA-free position to avoid clashing with the tRNA. Consequently the tRNA CCA end can bend and reach the aminoacylation active site. The tRNA 3′ region assumes two distinct conformations that allow A73 to be specifically recognized in different ways. One conformation is the canonical 'aminoacylation state.' The other conformation seems to be the 'intermediate state,' where the misaminoacylated 3′ end has partially relocated to the editing domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Larkin, D.C., Williams, A.M., Martinis, S.A. & Fox, G.E. Identification of essential domains for Escherichia coli tRNA(leu) aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 30, 2103–2113 (2002).
Asahara, H. et al. Recognition nucleotides of Escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr). J. Mol. Biol. 231, 219–229 (1993).
Small, I. et al. In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants: towards novel ways of influencing mitochondrial gene expression? EMBO J. 11, 1291–1296 (1992).
Breitschopf, K., Achsel, T., Busch, K. & Gross, H.J. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 23, 3633–3637 (1995).
Soma, A., Uchiyama, K., Sakamoto, T., Maeda, M. & Himeno, H. Unique recognition style of tRNA(Leu) by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 293, 1029–1038 (1999).
Yaremchuk, A., Kriklivyi, I., Tukalo, M. & Cusack, S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 21, 3829–3840 (2002).
Biou, V., Yaremchuk, A., Tukalo, M. & Cusack, S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science 263, 1404–1410 (1994).
Eriani, G., Delarue, M., Poch, O., Gangloff, J. & Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 (1990).
Rould, M.A., Perona, J.J., Soll, D. & Steitz, T.A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science 246, 1135–1142 (1989).
Ruff, M. et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science 252, 1682–1689 (1991).
Jakubowski, H. & Goldman, E. Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev. 56, 412–429 (1992).
Schmidt, E. & Schimmel, P. Residues in a class I tRNA synthetase which determine selectivity of amino acid recognition in the context of tRNA. Biochemistry 34, 11204–11210 (1995).
Lin, L., Hale, S.P. & Schimmel, P. Aminoacylation error correction. Nature 384, 33–34 (1996).
Nureki, O. et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 (1998).
Lincecum, T.L. et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell 11, 951–963 (2003).
Fukunaga, R., Fukai, S., Ishitani, R., Nureki, O. & Yokoyama, S. Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L–valine. J. Biol. Chem. 279, 8396–8402 (2004).
Fukai, S. et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell 103, 793–803 (2000).
Silvian, L.F., Wang, J. & Steitz, T.A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 (1999).
Chen, J.F., Guo, N.N., Li, T., Wang, E.D. & Wang, Y.L. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39, 6726–6731 (2000).
Baldwin, A.N. & Berg, P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem. 241, 839–845 (1966).
Hale, S.P., Auld, D.S., Schmidt, E. & Schimmel, P. Discrete determinants in transfer RNA for editing and aminoacylation. Science 276, 1250–1252 (1997).
Fukunaga, R. & Yokoyama, S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 346, 57–71 (2005).
Fukunaga, R. & Yokoyama, S. Structural basis for non-cognate amino acid discrimination by the valyl-tRNA synthetase editing domain. J. Biol. Chem. 280, 29937–29945 (2005).
Sekine, S., Nureki, O., Shimada, A., Vassylyev, D.G. & Yokoyama, S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol. 8, 203–206 (2001).
Delagoutte, B., Moras, D. & Cavarelli, J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 19, 5599–5610 (2000).
Hauenstein, S., Zhang, C.M., Hou, Y.M. & Perona, J.J. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 11, 1134–1141 (2004).
Fukai, S. et al. Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase. RNA 9, 100–111 (2003).
Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S. & Cusack, S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the editing conformation. Nat. Struct. Mol. Biol., advance online publication 11 September 2005 (10.1038/nsmb986)
Cusack, S., Yaremchuk, A. & Tukalo, M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 (2000).
Fukunaga, R., Ishitani, R., Nureki, O. & Yokoyama, S. Crystallization of leucyl-tRNA synthetase complexed with tRNALeu from the archaeon Pyrococcus horikoshii. Acta Crystallogr. F 61, 30–32 (2005).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. in Methods Enzymol. Vol. 276 (eds. Carter, C.W. Jr. & Sweet, R.M.) 307–326 (Academic Press, San Diego, USA, 1997).
Weeks, C.M. & Miller, R. Optimizing Shake-and-Bake for proteins. Acta Crystallogr. D Biol. Crystallogr. 55, 492–500 (1999).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Fukunaga, R. & Yokoyama, S. Crystallization and preliminary X-ray crystallographic study of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii. Acta Crystallogr. D Biol. Crystallogr. 60, 1916–1918 (2004).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
We thank M. Tukalo, A. Yaremchuk and S. Cusack for providing the coordinates of the T. thermophilus LeuRS–tRNALeu complex before publication and for helpful discussions. We thank R. Ishitani and O. Nureki for their help with crystal preparation and X-ray data collection. We also thank T. Sengoku, T. Yanagisawa, S. Sekine, M. Kawamoto, H. Sakai and M. Yamamoto for their help with data collection at BL41XU and BL26B1 in SPring-8. This work was supported by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, the RIKEN Structural Genomics/Proteomics Initiative (RSGI) and the National Project on Protein Structural and Functional Analyses, MEXT. R.F. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists and by SPring-8 Budding Researchers Support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Omit electron density maps of tRNA. (PDF 506 kb)
Supplementary Fig. 2
Two modes of discriminator recognition. (PDF 448 kb)
Rights and permissions
About this article
Cite this article
Fukunaga, R., Yokoyama, S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol 12, 915–922 (2005). https://doi.org/10.1038/nsmb985
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb985
This article is cited by
-
Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase
Nature Communications (2013)
-
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase
Nature Structural & Molecular Biology (2012)
-
Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation
Nature Structural & Molecular Biology (2009)
-
Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM
The EMBO Journal (2008)
-
Bringing order to translation: the contributions of transfer RNA anticodon‐domain modifications
EMBO reports (2008)