Abstract
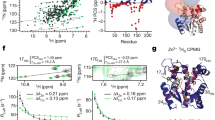
Although protein dynamics has been recognized as a potentially important contributor to enzyme catalysis, structural disorder is generally considered to reduce catalytic efficiency. This widely held assumption has recently been challenged by the finding that an engineered chorismate mutase combines high catalytic activity with the properties of a molten globule, a loosely packed and highly dynamic conformational ensemble. Taking advantage of the ordering observed upon ligand binding, we have now used NMR spectroscopy to characterize this enzyme in complex with a transition-state analog. The complex adopts a helix-bundle structure, as designed, but retains unprecedented flexibility on the millisecond timescale across its entire length. Moreover, pre–steady-state kinetics data show that binding occurs by an induced-fit mechanism on the same timescale as the enzymatic reaction, linking global conformational plasticity with efficient catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Karplus, M. Dynamics of proteins. Adv. Biophys. 18, 165–190 (1984).
Hammes, G.G. Multiple conformational changes in enzyme catalysis. Biochemistry 41, 8221–8228 (2002).
Boehr, D.D., Dyson, H.J. & Wright, P.E. An NMR perspective on enzyme dynamics. Chem. Rev. 106, 3055–3079 (2006).
Hammes-Schiffer, S. & Benkovic, S.J. Relating protein motion to catalysis. Annu. Rev. Biochem. 75, 519–541 (2006).
Olsson, M.H.M., Parson, W.W. & Warshel, A. Dynamical contributions to catalysis: critical tests of a popular hypothesis. Chem. Rev. 106, 1737–1756 (2006).
Dyson, H.J. & Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005).
Dunker, A.K., Brown, C.J., Lawson, J.D., Iakoucheva, L.M. & Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 41, 6573–6582 (2002).
MacBeath, G., Kast, P. & Hilvert, D. A small, thermostable, and monofunctional chorismate mutase from the archeon Methanococcus jannaschii. Biochemistry 37, 10062–10073 (1998).
MacBeath, G., Kast, P. & Hilvert, D. Redesigning enzyme topology by directed evolution. Science 279, 1958–1961 (1998).
Vamvaca, K., Vögeli, B., Kast, P., Pervushin, K. & Hilvert, D. An enzymatic molten globule: efficient coupling of folding and catalysis. Proc. Natl. Acad. Sci. USA 101, 12860–12864 (2004).
Bartlett, P.A. & Johnson, C.R. An inhibitor of chorismate mutase resembling the transition-state conformation. J. Am. Chem. Soc. 107, 7792–7793 (1985).
Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The X-plor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Lee, A.Y., Karplus, P.A., Ganem, B. & Clardy, J. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 117, 3627–3628 (1995).
Mittermaier, A. & Kay, L.E. New tools provide new insights in NMR studies of protein dynamics. Science 312, 224–228 (2006).
Palmer, A.G. & Massi, F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700–1719 (2006).
Lipari, G. & Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 (1982).
Jarymowycz, V.A. & Stone, M.J. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem. Rev. 106, 1624–1671 (2006).
Eletsky, A., Kienhöfer, A., Hilvert, D. & Pervushin, K. Investigation of ligand binding and protein dynamics in Bacillus subtilis chorismate mutase by transverse relaxation optimized spectroscopy-nuclear magnetic resonance. Biochemistry 44, 6788–6799 (2005).
Fersht, A. Structure and Mechanism in Protein Science (W. H. Freeman, New York, 1999).
Koshland, D.E. & Neet, K.E. The catalytic and regulatory properties of enzymes. Annu. Rev. Biochem. 37, 359–410 (1968).
Williams, D.H., Stephens, E., O'Brien, D.P. & Zhou, M. Understanding noncovalent interactions: ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem. Int. Ed. Engl. 43, 6596–6616 (2004).
Weber, G. Energetics of ligand binding to proteins. Adv. Protein Chem. 29, 1–83 (1975).
Masse, J.E. & Keller, R. AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J. Magn. Reson. 174, 133–151 (2005).
Herrmann, T., Güntert, P. & Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002).
Mumenthaler, C., Güntert, P., Braun, W. & Wüthrich, K. Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. J. Biomol. NMR 10, 351–362 (1997).
Schwieters, C.D., Kuszewski, J.J. & Clore, G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 (2006).
Güntert, P. Automated NMR protein structure calculation with CYANA. Meth. Mol. Biol. 278, 353–378 (2004).
Korzhnev, D.M., Billeter, M., Arseniev, A.S. & Orekhov, V.Y. NMR studies of Brownian tumbling and internal motions in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 38, 197–266 (2001).
Mandel, A.M., Akke, M. & Palmer, A.G. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 246, 144–163 (1995).
Cole, R. & Loria, J.P. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR 26, 203–213 (2003).
Acknowledgements
We are grateful to P. Anikeeva for carrying out preliminary NMR analysis, J. Beld for analytical ultracentrifugation studies, R. Kissner for technical assistance with stopped-flow experiments, and K. Woycechowsky for critical reading of the manuscript. This work was supported by the Schweizerischer Nationalfonds and the ETH Zurich. We dedicate this paper to the memory of D. Koshland.
Author information
Authors and Affiliations
Contributions
K.P. and D.H designed research; K.P. and B.V. did NMR experiments; K.V. did biochemical experiments; K.P., K.V., B.V. and D.H. analyzed data; K.P., K.V. and D.H. wrote the paper.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–3, Supplementary Methods (PDF 1322 kb)
Rights and permissions
About this article
Cite this article
Pervushin, K., Vamvaca, K., Vögeli, B. et al. Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol 14, 1202–1206 (2007). https://doi.org/10.1038/nsmb1325
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1325
This article is cited by
-
Mechanism of salt-induced activity enhancement of a marine-derived laccase, Lac15
European Biophysics Journal (2018)
-
Wild-type and molten globular chorismate mutase achieve comparable catalytic rates using very different enthalpy/entropy compensations
Science China Chemistry (2014)
-
Structure and function of a complex between chorismate mutase and DAHP synthase: efficiency boost for the junior partner
The EMBO Journal (2009)
-
Biological function in a non-native partially folded state of a protein
The EMBO Journal (2008)