Abstract
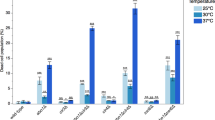
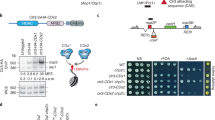
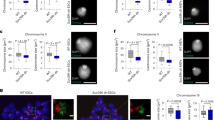
Heterochromatin causes epigenetic repression that can be transmitted through multiple cell divisions. However, the mechanisms underlying silencing and stability of heterochromatin are not fully understood. We show that heterochromatin differs from euchromatin in histone turnover and identify histone deacetylase (HDAC) Clr3 as a factor required for inhibiting histone turnover across heterochromatin domains in Schizosaccharomyces pombe. Loss of RNA-interference factors, Clr4 methyltransferase or HP1 proteins involved in HDAC localization causes increased histone turnover across pericentromeric domains. Clr3 also affects histone turnover at the silent mating-type region, where it can be recruited by alternative mechanisms acting in parallel to H3K9me–HP1. Notably, the JmjC-domain protein Epe1 promotes histone exchange, and loss of Epe1 suppresses both histone turnover and defects in heterochromatic silencing. Our results suggest that heterochromatic-silencing factors preclude histone turnover to promote silencing and inheritance of repressive chromatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003).
Grewal, S.I. & Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Richards, E.J. & Elgin, S.C. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108, 489–500 (2002).
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997).
Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D. & Grewal, S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bannister, A.J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Zofall, M. et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100 (2012).
Cam, H.P. et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37, 809–819 (2005).
Partridge, J.F., Borgstrom, B. & Allshire, R.C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14, 783–791 (2000).
Noma, K., Allis, C.D. & Grewal, S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155 (2001).
Djupedal, I. et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19, 2301–2306 (2005).
Kato, H. et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309, 467–469 (2005).
Volpe, T.A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Zhang, K., Mosch, K., Fischle, W. & Grewal, S.I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388 (2008).
Bayne, E.H. et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677 (2010).
Jia, S., Noma, K. & Grewal, S.I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 (2004).
Reyes-Turcu, F.E., Zhang, K., Zofall, M., Chen, E. & Grewal, S.I. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol. 18, 1132–1138 (2011).
Hall, I.M. et al. Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002).
Noma, K. et al. RITS acts in cis to promote RNA interference–mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36, 1174–1180 (2004).
Noma, K., Cam, H.P., Maraia, R.J. & Grewal, S.I. A role for TFIIIC transcription factor complex in genome organization. Cell 125, 859–872 (2006).
Schalch, T. et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol. Cell 34, 36–46 (2009).
Sadaie, M., Iida, T., Urano, T. & Nakayama, J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23, 3825–3835 (2004).
Yamada, T., Fischle, W., Sugiyama, T., Allis, C.D. & Grewal, S.I. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20, 173–185 (2005).
Sugiyama, T. et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504 (2007).
Motamedi, M.R. et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32, 778–790 (2008).
Fischer, T. et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. USA. 106, 8998–9003 (2009).
Yamane, K. et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41, 56–66 (2011).
Zofall, M. & Grewal, S.I. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692 (2006).
Isaac, S. et al. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175, 1549–1560 (2007).
Trewick, S.C., Minc, E., Antonelli, R., Urano, T. & Allshire, R.C. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 26, 4670–4682 (2007).
Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26 (2008).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Dion, M.F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Jamai, A., Imoberdorf, R.M. & Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007).
Rufiange, A., Jacques, P.E., Bhat, W., Robert, F. & Nourani, A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007).
Deal, R.B., Henikoff, J.G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010).
Garcia, J.F., Dumesic, P.A., Hartley, P.D., El-Samad, H. & Madhani, H.D. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24, 1758–1771 (2010).
Choi, E.S., Shin, J.A., Kim, H.S. & Jang, Y.K. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 33, 7102–7110 (2005).
Iacovoni, J.S., Russell, P. & Gaits, F. A new inducible protein expression system in fission yeast based on the glucose-repressed inv1 promoter. Gene 232, 53–58 (1999).
Lantermann, A.B. et al. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17, 251–257 (2010).
Thon, G., Cohen, A. & Klar, A.J. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138, 29–38 (1994).
Cam, H.P., Noma, K., Ebina, H., Levin, H.L. & Grewal, S.I. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451, 431–436 (2008).
Mito, Y., Henikoff, J.G. & Henikoff, S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 (2005).
Schwabish, M.A. & Struhl, K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 24, 10111–10117 (2004).
Venkatesh, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain–containing proteins. Nature 439, 811–816 (2006).
Braun, S. et al. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 (2011).
Ayoub, N. et al. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell Biol. 23, 4356–4370 (2003).
Grewal, S.I. & Klar, A.J. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146, 1221–1238 (1997).
Grewal, S.I., Bonaduce, M.J. & Klar, A.J. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150, 563–576 (1998).
Wirén, M. et al. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24, 2906–2918 (2005).
Kasten, M. et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23, 1348–1359 (2004).
Alfredsson-Timmins, J., Kristell, C., Henningson, F., Lyckman, S. & Bjerling, P. Reorganization of chromatin is an early response to nitrogen starvation in Schizosaccharomyces pombe. Chromosoma 118, 99–112 (2009).
Canzio, D. et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 (2011).
Kiely, C.M. et al. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol. Cell Biol. 31, 4193–4204 (2011).
Nicolas, E. et al. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14, 372–380 (2007).
Folco, H.D., Pidoux, A.L., Urano, T. & Allshire, R.C. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319, 94–97 (2008).
Acknowledgements
We are thankful to J. Barrowman for editing the manuscript, K. Zhang, K. Yamane and E. Luk for discussions, members of the Grewal laboratory for their help and P. Russell (Scripps Research Institute, La Jolla, California, USA) for the gift of pINV1 plasmid. This work is supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute. O.A. was supported by a European Molecular Biology Organization long-term post-doctoral fellowship.
Author information
Authors and Affiliations
Contributions
O.A. and S.I.S.G. designed research, O.A. performed all experiments, S.M. helped with microarray probe design, and O.A. and S.I.S.G. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 3058 kb)
Rights and permissions
About this article
Cite this article
Aygün, O., Mehta, S. & Grewal, S. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol 20, 547–554 (2013). https://doi.org/10.1038/nsmb.2565
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2565
This article is cited by
-
Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability
Nature Cell Biology (2023)
-
Histone deacetylation primes self-propagation of heterochromatin domains to promote epigenetic inheritance
Nature Structural & Molecular Biology (2022)
-
Mechanisms of chromatin-based epigenetic inheritance
Science China Life Sciences (2022)
-
Chromatin replication and epigenetic cell memory
Nature Cell Biology (2020)
-
Conserved protein Pir2ARS2 mediates gene repression through cryptic introns in lncRNAs
Nature Communications (2020)