Abstract
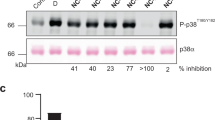
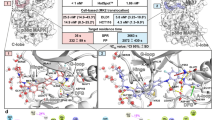
The quinazolinone and pyridol-pyrimidine classes of p38 MAP kinase inhibitors have a previously unseen degree of specificity for p38 over other MAP kinases. Comparison of the crystal structures of p38 bound to four different compounds shows that binding of the more specific molecules is characterized by a peptide flip between Met109 and Gly110. Gly110 is a residue specific to the α, β and γ isoforms of p38. The δ isoform and the other MAP kinases have bulkier residues in this position. These residues would likely make the peptide flip energetically unfavorable, thus explaining the selectivity of binding. To test this hypothesis, we constructed G110A and G110D mutants of p38 and measured the potency of several compounds against them. The results confirm that the selectivity of quinazolinones and pyridol-pyrimidines results from the presence of a glycine in position 110. This unique mode of binding may be exploited in the design of new p38 inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pearson, G. et al. Mitogen activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22, 153–183 (2001).
Lewis, T., Shapiro, P.S. & Ahn, N.G. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74, 49–139 (1998).
Davis, R.J. Transcriptional regulation by MAP kinases. Mol. Reprod. Dev. 42, 459–467 (1995).
Cobb, M.H. & Goldsmith, E.J. How MAP kinases are regulated. J. Biol. Chem. 270, 14843–14846 (1995).
Bokemeyer, D., Sorokin, A. & Dunn, M.J. Multiple intracellular MAP kinase signalling cascades. Kidney Int. 49, 1187–1198 (1996).
Derijard, B. et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025–1037 (1994).
Han, J., Lee, J.D., Bibbs, L. & Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265, 808–811 (1994).
Raingeaud, J. et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 270, 7420–7426 (1995).
Shapiro, L. & Dinarello, C.A. Osmotic regulation of cytokine synthesis in vitro. Proc. Natl. Acad. Sci. USA 92, 12230–12234 (1995).
Young, P.R. et al. Bicyclic imidazoles inhibit IL-1 and TNF production at the protein level. Agents Actions 39, C67–C69 (1993).
Guan, Z., Baier, L.D. & Morrison, A.R. p38 mitogen-activated protein kinase down-regulates nitric oxide and up-regulates prostaglandin E2 biosynthesis stimulated by interleukin-1β. J. Biol. Chem. 272, 8083–8089 (1997).
Raingeaud, J., Whitmarsh, A.J., Barrett, T., Derijard, B. & Davis, R.J. MKK3 and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16, 1247–1255 (1996).
Wang, X. & Ron, D. Stress-induced phosphorylation and activation of the transcription factor CHOP(GADD153) by p38 MAP kinase. Science 272, 1347–1349 (1996).
Han, J., Jiang, Y., Li, Z., Kravchenko, V.V. & Ulevitch, R.J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299 (1997).
Wu, T. et al. Involvement of 85-kd cytosolic phospholipase A(2) and cyclooxygenase-2 in the proliferation of human cholangiocarcinoma cells. Hepatology 36, 363–373 (2002).
Yamamori, T. et al. Relationship between p38 mitogen-activated protein kinase and small GTPase Rac for the activation of NADPH oxidase in bovine neutrophils. Biochem. Biophys. Res. Comm. 293, 1571–1578 (2002).
Revesz, L. et al. SAR of 4-hydroxypiperidine and hydroxyalkayl substituted heterocycles as novel p38 MAP kinase inhibitors. Bioorg. Med. Chem. Lett. 10, 1261–1264 (2000).
Wilson, K.P. et al. The structural basis for the specificity of pyridinylimidazole inhibitors of p38 MAP kinase. Chem. Biol. 4, 423–431 (1997).
Lisnock, J.M. et al. Molecular basis for p38 protein kinase inhibitor specificity. Biochemistry 37, 16573–16581 (1998).
Wang, Z. et al. Structural basis of inhibitor selectivity in MAP kinases. Structure 6, 1117–1128 (1998).
Tong, L. et al. A highly specific inhibitor of human p38 MAP kinase binds in the ATP binding pocket. Nat. Struct. Biol. 4, 311–316 (1997).
Natarajan, S.R. et al. p38 MAP kinase inhibitors part 1: design and development of a new class of potent and highly selective inhibitors based on 3,4-dihydropyrido[3,2-d]pyrimidone scaffold. Bioorg. Med. Chem. Lett. 13, 273–276 (2003).
Hunt, J.A. et al. p38 inhibitors: piperidine- and 4-aminopiperidine-substituted naphthyridinones, quinolinones, and dihydroquinazolinones. Bioorg. Med. Chem. Lett. 13, 467–470 (2003).
Stelmach, J.E. et al. Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase. Bioorg. Med. Chem. Lett. 13, 277–280 (2003).
Liverton, N.J. et al. Design and synthesis of potent, selective, and orally bioavailable tetrasubstituted imidazole inhibitors of p38 mitogen-activated protein kinase. J. Med. Chem. 42, 2180–2190 (1999).
Boehm, J.C. & Adams, J.L. New inhibitors of p38 kinase. Expert Opin. Ther. Pat. 10, 25–37 (2000).
Toledo, L.M., Lydon, N.B. & Elbaum, D. The structure-based design of ATP-site directed protein kinases inhibitors. Curr. Med. Chem. 6, 775–805 (1999).
Nar, H., Messerschmidt, A., Huber, R., van de Kamp, M. & Canters, G.W. Crystal structure analysis of oxidized Pseudomonas aeruginosa azurin at pH 5.5 and pH 9.0. A pH-induced conformational transition involves a peptide bond flip. J. Mol. Biol. 221, 765–772 (1991).
Hayward, S. Peptide-plane flipping in proteins. Protein Sci. 10, 2219–2227 (2001).
Gunasekaran, K., Gomathi, L., Ramakrishnan, C., Chandrasekhar, J. & Balaram, P. Conformational interconversions in peptide β-turns: analysis of turns in proteins and computational estimates of barriers. J. Mol. Biol. 284, 1505–1516 (1998).
Pargellis, C. et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 9, 268–272 (2002).
Howard, A.J. Data processing in macromolecular crystallography. In Crystallographic Computing 7: Proceedings from the Macromolecular Crystallographic Computing School, 1996 (eds. Bourne, E.P.E & Watenpaugh, K.D.) (Oxford Univ. Press, Oxford, UK, 2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Wilson, K.P. et al. Crystal structure of p38 mitogen-activated protein kinase. J. Biol. Chem. 271, 27696–27700 (1996).
Read, R.J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140–149 (1986).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, A.T., Zuo, J.Y., Cowan, S.W. and Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Laskoswski, R.A., MacArthur, M.W., Moss, S.D. & Thornton, J.M. PROCHECK: a programme to check the stereochemical quality of protein structure coordinates. J. Appl. Crystallogr. 26, 283–291 (1993).
Carson, M. Ribbons. Methods Enzymol. 277, 493–505 (1997).
Acknowledgements
We thank the staff at the facilities at the IMCA-CAT for help during data collection. The facilities at IMCA-CAT are supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Illinois Institute of Technology (IIT), executed through IIT's Center for Synchrotron Radiation Research and Instrumentation. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Fitzgerald, C., Patel, S., Becker, J. et al. Structural basis for p38α MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity. Nat Struct Mol Biol 10, 764–769 (2003). https://doi.org/10.1038/nsb949
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb949
This article is cited by
-
The pyridazine heterocycle in molecular recognition and drug discovery
Medicinal Chemistry Research (2023)
-
Kinase inhibition in autoimmunity and inflammation
Nature Reviews Drug Discovery (2021)
-
Data driven polypharmacological drug design for lung cancer: analyses for targeting ALK, MET, and EGFR
Journal of Cheminformatics (2017)
-
Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor
Nature Chemical Biology (2012)
-
Virtual screening using a conformationally flexible target protein: models for ligand binding to p38α MAPK
Journal of Computer-Aided Molecular Design (2012)