Abstract
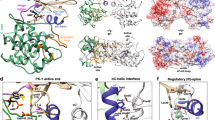
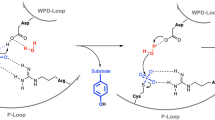
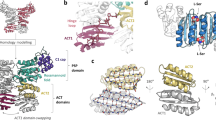
A family of eukaryotic-like Ser/Thr protein kinases occurs in bacteria, but little is known about the structures and functions of these proteins. Here we characterize PknB, a transmembrane signaling kinase from Mycobacterium tuberculosis. The intracellular PknB kinase domain is active autonomously, and the active enzyme is phosphorylated on residues homologous to regulatory phospho-acceptors in eukaryotic Ser/Thr kinases. The crystal structure of the PknB kinase domain in complex with an ATP analog reveals the active conformation. The predicted fold of the PknB extracellular domain matches the proposed targeting domain of penicillin-binding protein 2x. The structural and chemical similarities of PknB to metazoan homologs support a universal activation mechanism of Ser/Thr protein kinases in prokaryotes and eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cheek, S., Zhang, H., & Grishin, N.V. Sequence and structure classification of kinases. J. Mol. Biol. 320, 855–881 (2002).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Cole, S.T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (2000).
Av-Gay, Y. & Everett, M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8, 238–244 (2000).
Andersen, P. TB vaccines: progress and problems. Trends Immunol. 22, 160–168 (2001).
Ono-Saito, N., Niki, I. & Hidaka, H. H-series protein kinase inhibitors and potential clinical applications. Pharmacol. Ther. 82, 123–131 (1999).
Drews, S.J., Hung, F. & Av-Gay, Y. A protein kinase inhibitor as an antimicrobial agent. FEMS Microbiol. Lett. 205, 369–374 (2001).
Chaba, R., Raje, M. & Chakrobarti, P.K. Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur. J. Biochem. 269, 1078–1085 (2002).
Av-Gay, Y., Jamil, S. & Drews, S.J. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infection Immunol. 67, 5676–5682 (1999).
Koul. A. et al. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology 147, 2307–2314 (2001).
Huse, M. & J. Kuriyan . The conformational plasticity of protein kinases. Cell 109, 275–282 (2002).
Niefind, K., Guerra, B., Ermakowa, I. & Issinger, O.G. Crystal structure of human protein kinase Ck2: Insights into basic properties of the Ck2 holoenzyme. Embo J. 20, 5320–5331 (2001).
Madhusudan, Akamine, P., Xuong, N.-H. & Taylor, S.S. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat. Struct. Biol. 9, 273–277 (2002).
Goldberg J., Nairn, A.C., & Kuriyan, J. Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell 84, 875–887 (1996).
Kelley, L.A., MacCallum, R.M. & Sternberg, M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520 (2000).
Gordon, E., Mouz, N., Duee, E. & Dideberg, O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299, 477–485 (2000).
Marchler-Bauer, A. et al. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30, 281–283 (2002).
Yeats, C. Finn, R.D. & Bateman, A. The PASTA domain: a β-lactam binding domain. Trends Biochem. Sci. 9, 438–40 (2002).
Schulze-Gahmen, U., et al. Multiple modes of ligand recognition: crystal structures of cyclin-dependent protein kinase 2 in complex with ATP and two inhibitors, olomoucine and isopentenyladenine. Proteins 22, 378–391 (1995).
Xie, X. et al. Crystal structure of JNK3: a kinase implicated in neuronal apoptosis. Structure 6, 983–991 (1998).
Zhang F., Strand A., Robbins D., Cobb M.H., & Goldsmith E.J. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature 367, 704–711 (1994).
Xu, W., Doshi, A., Lei, M., Eck, M.J. & Harrison, S.C. Crystal structures of c-Src reveal new features of its autoinhibitory mechanism. Mol. Cell 3, 629–638 (1999).
Leonard, C.J., Aravind, L. & Koonin, E.V. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 8, 1038–1047 (1998).
Han, G. & Zhang, C.-C. On the origin of Ser/Thr kinases in a prokaryote. FEMS Microbiol. Lett. 200, 79–84 (2001).
Young, M.A., Gonfloni, S., Superti-Furga, G., Roux, B. & Kuriyan, J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell 105, 115–126 (2001).
Juris S.J., Rudolph, A.E., Huddler, D., Orth, K. & Dixon, J.E. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc. Natl. Acad. Sci. USA 15, 9431–9436 (2000).
Havlir, D.V. et al. Human immune response to Mycobacterium tuberculosis antigens. Infection Immunol. 59, 665–670 (1991).
Durocher, D. & Jackson, S.P. The FHA domain. FEBS Let. 513, 58–66 (2002).
Marcotte, E.M. et al. Detecting protein function and protein–protein interactions from genome sequences. Science 285, 751–753 (1999).
Pham, K., LaForge, K.S. & Kreek, M.J. Sticky-end PCR: new method for subcloning. Biotechniques 25, 206–208 (1998).
Bienvenut, W.V. et al. Matrix-assisted laser desorption/ionization-tandem mass spectrometry with high resolution and sensitivity for identification and characterization of proteins. Proteomics 2, 868–876 (2002).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Jones T.A., Zou J.Y., Cowan S.W. & Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Brunger A.T., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Laskowski, R.A, MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 26, 283–291 (1993).
Chang, C.-I., Xu, B., Akella, R. Cobb, M.H. & Goldsmith, E.J. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9, 1241–1249 (2002).
Acknowledgements
We thank D.S. King for protein molecular weight measurements, K.F. Medzihradszky for LC/MS/MS measurements, J. Holton and E. Skordalakes for help with structure determination, C.A. Settineri for help with the deconvoluted protein mass spectrum, and L. Gay and M. Good for stimulating discussions. We are indebted to J. Kuriyan for encouragement and for pointing out the importance of the regulatory interaction sites in PknB-I. K. Strenge and S. Shieh provided technical support. We are indebted to T. Terwilliger and the TB Structural Genomics Consortium for support. The work was supported by a grant from the NIH. The Advanced Light Source Beamline 8.3.1 was funded by the NSF, the University of California and Henry Wheeler.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Young, T., Delagoutte, B., Endrizzi, J. et al. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Mol Biol 10, 168–174 (2003). https://doi.org/10.1038/nsb897
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb897
This article is cited by
-
Spectroscopic, quantum chemical, ADMET and molecular docking studies of echinatin: a prospective tuberculosis drug
Research on Chemical Intermediates (2022)
-
Are antibacterial effects of non-antibiotic drugs random or purposeful because of a common evolutionary origin of bacterial and mammalian targets?
Infection (2021)
-
Potential therapeutic approaches for a sleeping pathogen: tuberculosis a case for bioinorganic chemistry
JBIC Journal of Biological Inorganic Chemistry (2020)
-
Novel mechanistic insights into physiological signaling pathways mediated by mycobacterial Ser/Thr protein kinases
Genes & Immunity (2019)
-
LipidII interaction with specific residues of Mycobacterium tuberculosis PknB extracytoplasmic domain governs its optimal activation
Nature Communications (2019)