Abstract
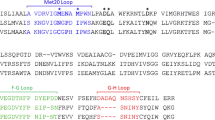
Bacteria expressing R67-plasmid encoded dihydrofolate reductase (R67 DHFR) exhibit high-level resistance to the antibiotic trimethoprim. Native R67 DHFR is a 34,000 Mr homotetramer which exists in equilibrium with an inactive dimeric form. The structure of native R67 DHFR has now been solved at 1.7 Å resolution and is unrelated to that of chromosomal DHFR. Homotetrameric R67 DHFR has an unusual pore, 25 Å in length, passing through the middle of the molecule. Two folate molecules bind asymmetrically within the pore indicating that the enzyme's active site consists of symmetry related binding surfaces from all four identical units.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Kraut, J. & Matthews, D.A. in Active sites of Enzymes (eds Jurnak, F.A. & McPherson, A.) 3, 1–71 (John Wiley & sons, New York, 1987).
Fleming, M.P., Datta, N. & Gruneberg, R.N. Trimethoprim resistance determined by R factors. Br. med. J. 1, 726–728 (1972).
Pattishall, K.H. et al. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J. biol. Chem. 252, 2319–2323 (1977).
Smith, S.L. et al. R plasmid dihydrofolate reductase with subunit structure. J. biol. Chem. 254, 6222–6225 (1979).
Stone, D. & Smith, S.L. The amino acid sequence of the trimethoprim-resistant dihydrofolate reductase specified in Escherichia coli by R-plasmid R67. J. biol. Chem. 254, 10857–10861 (1979).
Nichols, R. et al. Titration of histidine 62 in R67 dihydrofolate reductase is linked to a tetramer ⟺ two-dimer equilibrium. Biochemistry 32, 1695–1706 (1993).
Matthews, D.A. et al. Crystal structure of a novel trimethoprim-resistant dihydrofolate reductase specified in Escherichia coli by R-plasmid R67. Biochemistry 25, 4194–4204 (1986).
Reece, L.J., Nichols, R., Ogden, R.C. & Howell, E.E. Construction of a synthetic gene for an R-plasmid encoded dihydrofolate reductase and studies on the role of the N-terminus in the protein. Biochemistry 30, 10895–10904 (1991).
Janin, J., Miller, S. & Chothia, C. Surface, subunit interfaces and interior of oligomeric proteins. J. molec. Biol. 204, 155–164 (1988).
Braig, K. et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371, 578–586 (1994).
Bennett, M.J., Choe, S. & Eisenberg, D. Domain swapping: entangling alliances between proteins. Proc. natn. Acad. Sci. U.S.A. 91, 3127–3131 (1994).
Matthews, B.W. & Bernhard, S.A. Structure and symmetry of oligomeric enzymes. A. Rev. Biophys. Bioengng. 2, 257–317 (1973).
Navia, M.A. et al. Three-dimensional structure of aspartyl protease from human immuno deficiency virus HIV-1. Nature 337, 615–620 (1989).
Holland, J.C. et al. in Proc 10th Intl. symp. chem. biol. pteridines (eds Ayling, J. E. et al.) 493–498 (Plenum, New York, 1993).
Brito, R.M.M. et al. Characterization and stereochemistry of cofactor oxidation by a type II dihydrofolate reductase. Biochemistry 29, 9825–9831 (1990).
Morrison, J.F. & Sneddon, M.K. in Chemistry and Biology of Pteridines (eds Curtius, H.-Ch., Ghisla, S. & Bau, N.) 728–733 (Walter de Gruyter, Berlin, 1990).
Bystroff, C., Oatley, S.J. & Kraut, J. Crystal structures of Escherichia coli dihydrofolate reductase: The NADP+ holoenzyme and the folate. NADP+ ternary complex. Substrate binding and a model for the transition state. Biochemistry 29, 3263–3277 (1990).
Brito, R.M.M., Rudolph, F.B. & Rosevear, P.R. Conformation of NADP+ bound to a type II dihydrofolate reductase. Biochemistry 30, 1461–1469 (1991).
Smith, S.L. & Burchall, J. α-Pyridine nucleotides as substrates for a plasmid-specified dihydrofolate reductase. Proc. natn. Acad. Sci. U.S.A. 80, 4619–4623 (1983).
Borchert, T.V., Mathieu, M. & Zeelen, J.P. The crystal structure of human Csk SH3: Structural diversity near the RT-Src and n-Src loop. FEBS Lett. 341, 79–85 (1994).
Kohda, D. et al. Solution structure and ligand-binding site of the carboxy-terminal SH3 domain of GRB2. Structure 2, 1029–1040 (1994).
Musacchio, A., Saraste, M. & Wilmanns, M. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nature struct. Biol. 1, 546–551 (1994).
Wilson, K.P. et al. Escherichia coli biotin holoenzyme synthetase/ bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. natn. Acad. Sci. U.S.A. 89, 9257–9261 (1992).
Falzone, C.J. et al. Three-dimensional solution structure of PsaE from the cyanobacterium synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains. Biochemistry 33, 6052–6062 (1994).
Baumann, H. et al. Solution structure and DNA-binding properties of a thermostable protein from the archaeon sulfolobus solfataricus. Nature struct. Biol. 1, 808–819 (1994).
Jencks, W.P. Catalysis in Chem. and Enzym., (McGraw-Hill, New York; 1975).
Matthews, B.W. Solvent content of protein crystals. J. molec. Biol. 33, 491–497 (1968).
Hamlin, R. Multiwire area X-ray diffractometers. Meths Enzym. 114A, 416–452 (1985).
Howard, A.J., Nielsen, C., Xuong, N.-h. Software for a diffractometer with multiwire area detector. Meths. Enzym. 114, 452–472 (1985).
Eleanor, D. et al. A programme system from Oxford, UK.
Terwilliger, T.C. & Eisenberg, D. Unbiased three-dimensional refinement of heavy-atom parameters by correlation of origin-removed patterson functions. Acta crystallogr. A39, 813–817 (1983).
Wang, B.-C. Resolution of phase ambiguity in macromolecular crystallography. Meths Enzym. 115B, 90–112 (1985).
Cambillau, C. & Horjales, E. TOM: a FRODO subpackage for protein-ligand fitting with interactive energy minimization. J. molec. Graph. 5, 174–177 (1987).
Brunger, A.T. X-PLOR vers. 3.1 Manual: A system for X-ray Crystallography & NMR (1992). Yale University, New Haven, CT 06511, U.S.A.
Tronrud, D.E., Ten Eyck, L.F. & Matthews, B.W. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta crystallogr. A43, 489–501 (1987).
Laskowski, R.A., MacArthur, M.W., Moss, D.S., & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. appl. Crystallogr. 26, 283–291 (1993).
Ramachandran, G.N. & Sasisekharan, V. Conformation of polypeptides and proteins. Adv. prot. Chem. 23, 283–438 (1968).
Luzzati, P.V. Traitement statistique des erreurs dans la determination des structures cristallines. Acta crystallogr. 5, 802–810 (1952).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. appl. Crystallogr. 24, 946–950 (1991).
Insight II User Guide, Vers. 2.3.9 (1994), Biosym Technologies, Inc., San Diego.
Connolly, M.L. Analytical molecular surface calculation. J. appl. Crystallogr. 16, 548–558 (1983).
Bolin, J.T. et al. Crystal structures of Escherichia coli and Lactobacillus casei dihydro-folate reductase refined a 1.7 Å resolution. I. General features and binding of methotrexate. J. biol. Chem. 257, 13650–13662 (1982).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Narayana, N., Matthews, D., Howell, E. et al. A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site. Nat Struct Mol Biol 2, 1018–1025 (1995). https://doi.org/10.1038/nsb1195-1018
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb1195-1018
This article is cited by
-
Multiple ligand-binding modes in bacterial R67 dihydrofolate reductase
Journal of Computer-Aided Molecular Design (2005)