Abstract
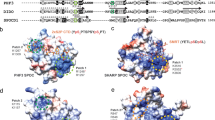
The basic region leucine zipper (bZIP) proteins form one of the largest families of transcription factors in eukaryotic cells. Despite relatively high homology between the amino acid sequences of the bZIP motifs, these proteins recognize diverse DNA sequences. Here we report the 2.0 Å resolution crystal structure of the bZIP motif of one such transcription factor, PAP1, a fission yeast AP-1-like transcription factor that binds DNA containing the novel consensus sequence TTACGTAA. The structure reveals how the Pap1-specific residues of the bZIP basic region recognize the target sequence and shows that the side chain of the invariant Asn in the bZIP motif adopts an alternative conformation in Pap1. This conformation, which is stabilized by a Pap1-specific residue and its associated water molecule, recognizes a different base in the target sequence from that in other bZIP subfamilies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Pabo, C.O. et al. Science 247,1210–1213 (1990).
Wolberger, C., Vershon, A.K., Liu, B., Johnson, A.D. & Pabo, C.O. Cell 67, 517–528 (1991).
Pomerantz, J.L. & Sharp, P.A. Biochemistry 33, 10851–10858 (1994).
Choo, Y. & Klug, A. Curr. Opin. Struct. Biol. 7, 117–125 (1997).
Toda, T. et al. Mol. Cell. Biol. 12, 5474–5484 (1992).
Shimanuki, M., Saka, Y., Yanagida, M. & Toda, T. J. Cell Sci. 108, 569–579 (1995).
Toone, W.M. et al. Genes Dev. 12, 1453–1463 (1998).
Fernandes, L., Rodrigues-Pousada, C. & Struhl, K. Mol. Cell. Biol. 17, 6982–6993 (1997).
Benbrook, D.M. & Jones, N.C. Nucleic Acids Res. 22, 1463–1469 (1994).
Ellenberger, T.E., Brandl, C.J., Struhl, K. & Harrison, S.C. Cell 71, 1223–1237 (1992).
Koning, P. & Richmond, T.J. J. Mol. Biol. 233, 139–154 (1993).
Glover, J.N.M. & Harrison, S.C. Nature 373, 257–261 (1995).
Chen, L., Glover, J.N.M., Hogan, P.G., Rao, A. & Harrison, S.C. Nature 392, 42–48 (1998).
Xu, W., Rould, M.A., Jun, S., Desplan, C. & Pabo, C.O. Cell 80, 639–650 (1995).
Schwartz, T., Rould, M.A., Lowenhaupt, K., Herbert, A. & Rich, A. Science 284, 1841–1845 (1999).
Keller, W., Konig, P. & Richmond, T.J. J. Mol. Biol. 254, 657–667 (1995).
Baxevanis, A.D. & Vinson, C.R. Curr. Opin. Gen. Dev. 3, 278–285 (1993).
Ferre-D'Amare, A.D., Prendergast, G.C., Ziff, E.B. & Burley, S.K. Nature 363, 38–44 (1993).
Brownlie, P., et al. Structure 5, 509–520 (1997).
Ferre-D'Amare, A.D., Pognonec, P., Roeder, R.G. & Burley, S.K. EMBO J. 13, 180–189 (1994).
Shimizu, T. et al. EMBO. J. 16, 4689–4697 (1997).
Koldin, B., Suckow, M., Seydel, A., Wilcken-Bergmann, B. & Muller-Hill, B. Nucleic Acids Res. 23, 4162–4169 (1995)
Vinson, C.R., Sigler, P.B. & McKnight, S.L. Science 246, 911–916 (1989).
Johnson, P.F. (1993). Mol. Cell. Biol. 13, 6919–6930 (1993).
Benbrook, D.M. & Jones, N.C. Nucleic Acids Res. 22, 1463–1469 (1994).
Fujii, Y. et al. Acta Crystallogr. D 54, 1014–1016 (1998).
Otwinowski, Z. & Monor, W. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. Acta Crystallogr. D 50, 760–763 (1994).
Cowtan, K. & Main, P. Acta Crystallogr. D 52, 43–48 (1996).
Jones, T.A., Zou. J.-Y., Cowan, S.W. & Kjeldgaad, M. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
This work was supported by Grants in Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to T.H.). We thank Y. Kyogoku for support at the early stage of the project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujii, Y., Shimizu, T., Toda, T. et al. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Mol Biol 7, 889–893 (2000). https://doi.org/10.1038/82822
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/82822
This article is cited by
-
bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum
Microbial Cell Factories (2022)
-
Specifically bound BZIP transcription factors modulate DNA supercoiling transitions
Scientific Reports (2020)
-
Molecular mechanisms of the protein-protein interaction–regulated binding specificity of basic-region leucine zipper transcription factors
Journal of Molecular Modeling (2019)
-
Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer
Scientific Reports (2018)
-
Genome-wide characterization of the basic leucine zipper transcription factors in Camellia sinensis
Tree Genetics & Genomes (2018)