Abstract
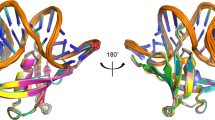
Many biochemical processes, including DNA packing, maintenance and control, rely on non-sequence specific protein–DNA interactions. Nonspecific DNA-binding proteins have evolved to tolerate a wide range of DNA sequences, yet bind with a respectable affinity. The nonspecific binding requirement is in contrast to that imposed on, for example, transcription factors and implies a different structural basis for the biomolecular recognition process. To address this issue, and the mechanism for archaeal DNA packing, we determined the structure of the Sso7d protein from Sulfolobus solfataricus in complex with DNA. Sso7d binds DNA by placing a triple-stranded β-sheet across the DNA minor groove. The protein is anchored in this position by the insertion of hydrogen bond-donating side chains into the groove and additionally stabilized by electrostatic and non-polar interactions with the DNA backbone. This structure explains how strong binding can be achieved independent of DNA sequence. Sso7d binding also distorts the DNA conformation and introduces significant unwinding of the helix. This effect suggests a mechanism for DNA packing in Sulfolobus based on negative DNA supercoiling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Travers, A.A., Ner, S.S. & Churchill, M.E.A. DNA chaperones: A solution to a persistence problem. Cell 77, 167–69 ( 1994).
Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251 –260 (1997).
Starich, M.R., Sandman, K., Reeve, J.N. & Summers, M.F. NMR structure of HMfB from the hyperthermophile Methanothermus fervidus confirms that this archaeal protein is a histone. J. Mol. Biol. 25, 187–203 (1996).
Shioda, M.B., Sugimori, K., Shiroya, T. & Takayanagi, S. Nucleosomelike structures associated with chromosomes of the archaebacterium Halobacterium salinarium. J. Bacteriol. 171, 4514– 4517 (1989).
Bohrmann, B. et al. Localization of histone-like proteins in thermophilic archaea by immunogold microscopy. J. Struct. Biol. 112, 70– 78 (1994).
Thomm, M., Stetter, K.O. & Zillig, W. Histone-like proteins in eu- and archaebacteria. Zentralbl. Bakeriol. Mikrobiol. Hyg., I. Abt. Orig. C 3, 128– 139 (1982).
Dijk, J. & Reinhardt, R. The structure of DNA-binding proteins from eu and archaebacteria. in Bacterial Chromatin (eds Gualerzi, C.O. & Pon, C.L.) 185–218 (Springer-Verlag, Berlin, 1986).
Choli, T., Henning, P., Wittmann-Liebold, B. & Reinhardt, R. Isolation, characterization, and microsequence analysis of a small basic methylated DNA-binding protein from the archaebacterium Sulfolobus solfataricus. Biochim. Biophys. Acta 950, 193– 203 (1988).
Baumann, H., Knapp, S., Lundbäck, T., Ladenstein, R. & Härd, T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nature Struct. Biol. 1, 808–819 (1994).
Lundbäck, T. & Härd, T. Salt dependence of the free energy, enthalpy and entropy of nonsequence specific DNA binding. J. Phys. Chem. 100, 17690–17695 ( 1996).
McAfee, J.G., Edmondson, S.P., Zegar, I. & Shriver, J.W. Equilibrium DNA Binding of Sac7d Protein from the Hyperthermophile Sulfolobus acidocaldarius : fluorescence and circular dichroism studies. Biochemistry 35, 4034–4045 ( 1996).
Lundbäck, T., Hansson, H., Knapp, S., Ladenstein, R. & Härd, T. Thermodynamic characterization of nonsequence specific DNA-binding by the Sso7d protein from Sulfolobus solfataricus. J. Mol. Biol. 276, 775–786 (1998).
Brunger, A. X-PLOR version 3.1: A system for X-ray crystallography and NMR, (Yale University Press, New Haven, Connecticut; 1992).
Robinson, H. et al. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392, 202–205 ( 1998).
Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12, 861– 867 (1993).
Baumann, H., Knapp, S., Karshikoff, A., Ladenstein, R. & Härd, T. DNA-binding surface of the Sso7d protein from Sulfolobus solfataricus. J. Mol. Biol. 247, 840 –846 (1995).
Guagliardi, A., Cerchia, L., Camardella, L., Rossi, M. & Bartolucci, S. DBF (disulfide bond-forming) enzyme from the hyperthermophilic archaebacterium Sulfolobus solfataricus behaves like a molecular chaperone. Biocatalysis 11, 181–190 (1994).
Saraste, M., Sibbald, P.R. & Wittinghofer, A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430– 434 (1990).
Berglund, H., Baumann, H., Knapp, S., Ladenstein, R. & Härd, T. Flexibility of an arginine side chain at a DNA-protein interface. J. Am. Chem. Soc. 117, 12883– 12884 (1995).
Knapp, S. et al. Thermal unfolding of the DNA-binding protein Sso7d from the hyperthermophile Sulfolobus solfataricus. J. Mol. Biol. 264, 1132–1144 (1997).
Rhodes, D. & Klug, A. Helical periodicity of DNA determined by enzyme digestion. Nature 286, 573– 578 (1980).
Depew, R.E. & Wang, J.C. Conformational fluctuations of DNA helix. Proc. Natl. Acad. Sci. USA 72, 4275 –4279 (1975).
López-Garcia, P., Knapp, S., Ladenstein, R. & Forterre, P. In vitro DNA binding of the archaeal protein Sso7d induces negative supercoiling at temperatures typical for thermophilic growth. Nucleic Acids. Res., in the press (1998).
Macura, A. & Ernst, R.R. Elucidation of crossrelaxation in liquids by 2D NMR spectroscopy. Mol. Phys. 41, 95–117 (1980).
Kay, L.E. Pulsed field gradient multi-dimensional NMR methods for the study of protein structure and dynamics in solution. Prog. Biophys. Mol. Biol. 63, 277–299 (1995).
Slijper, M., Kaptein, R. & Boelens, R. Simultaneous 13C and 15N isotope editing of biomolecular complexes. Application to a mutant lac repressor headpiece DNA complex. J. Magn. Reson. B 111, 199– 203 (1996).
Wuthrich, K. NMR of protein and nucleic acids. (John Wiley & Sons, New York; 1986).
Vuister, G.W. & Bax, A. Quantitative J correlation: a new approach for measuring homonuclear threebond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115 , 7772–7777 (1993).
Archer, S.J., Ikura, M., Torchia, D.A. & Bax, A. An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson. 95, 636–641 (1991).
Vuister, G.W. & Bax, A. Measurement of two- and three-bond proton to methyl-carbon J couplings in proteins uniformly enriched with 13C. J. Magn. Reson. B 102, 228– 231 (1993).
Kim, S.-G., Lin, L.-J. & Read, B.R. Determination of nucleic acid backbone conformation by 1H NMR . Biochemistry 31, 3564– 3574 (1992).
Omichinski, J.G., Pedone, P.V., Felsenfeld, G., Gronenborn, A.M. & Clore, G.M. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nature Struct. Biol. 4, 122–132 (1997).
Nilges, M. A calculations strategy for the structure determination of symmetric dimers by 1H NMR. Proteins 17, 297–309 (1993).
Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biol. NMR 8, 477–486 (1996).
Lavery, R. & Skelnar, H. Defining the structure of irregular nucleid acids: conventions and principles. J. Biomol. Struct. Dyn. 6, 655–667 ( 1989).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures . J. Appl. Crystallogr. 24, 946– 950 (1991).
Bacon, D.J. & Anderson, W.F. A fast algorithm for rendering space-filling molecule pictures. Mol. Graphics 6, 219–220 (1988).
Acknowledgements
This work was supported by the Swedish Natural Sciences Research Council (NFR), the Magn. Bervall Foundation and by the EC project Biotechnology of Extremophiles. We thank Mats Wikström and Toshi Nishida at Pharmacia & Upjohn for providing measurement time on an 800 MHz NMR spectrometer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Agback, P., Baumann, H., Knapp, S. et al. Architecture of nonspecific protein–DNA interactions in the Sso7d–DNA complex. Nat Struct Mol Biol 5, 579–584 (1998). https://doi.org/10.1038/836
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/836
This article is cited by
-
Programmable Assembly of DNA-protein Hybrid Structures
Chemical Research in Chinese Universities (2020)
-
The archaeal “7 kDa DNA-binding” proteins: extended characterization of an old gifted family
Scientific Reports (2016)
-
Biochemical characterization of DNA-binding proteins from Pyrobaculum aerophilum and Aeropyrum pernix
Extremophiles (2008)
-
Evolutionary conservation and DNA binding properties of the Ssh7 proteins fromSulfolobus shibatae
Science in China Series C: Life Sciences (2002)
-
The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA
Nature Structural Biology (1998)