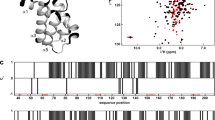
Abstract
Both prokaryotes and eukaryotes regulate transcription through mechanisms that suppress termination signals. An antitermination mechanism was first characterized in bacteriophage λ. Bacteria have analogous machinery that regulates ribosomal RNA transcription and employs host factors, called the N-utilizing (where N stands for the phage λ N protein) substances (Nus), NusA, NusB, NusE and NusG. Here we report the crystal structure of NusB from Mycobacterium tuberculosis, the bacterium that causes tuberculosis in humans. This molecule shares a similar tertiary structure with the related Escherichia coli protein but adopts a different quaternary organization. We show that, unlike the E. coli homolog, M. tuberculosis NusB is dimeric both in solution and in the crystal. These data help provide a framework for understanding the structural and biological function of NusB in the prokaryotic transcriptional antitermination complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Friedman, D.I. & Court, D.L. Mol. Microbiol. 18, 191–200 (1995).
Keener, J. & Nomura, M. In Cellular and molecular biology (eds Neidhardt, F.C. et al.) 822–848 (American Society for Microbiology, Washington, DC; 1996).
Zellers, M. & Squires, C.L. Mol. Microbiol. 32, 1296–1304 (1999).
Taura, T., Ueguchi, C. & Shiba, K, Ito, K. Mol. Gen. Genet 234, 429–432 (1992).
Cole, S.T. et al. Nature 393, 537–544 (1998).
Ji, Y. & Colston, M.J, Cox, R.A. Microbiol. 140, 123–132 (1994).
Ji, Y. & Colston, M. J, Cox, R.A. Microbiol. 140, 2829–2840 (1994).
Bremer, H. & Dennis, P.P. In Cellular and molecular biology. (eds Neidhardt, F.C. et al.) 1527–1542 (American Society for Microbiology, Washington, DC; 1987).
Bercovier, H., Kafri, O. & Sela, S. Biochem. Biophys. Res. Comm. 136, 1136–1141 (1986)
Huenges, M. et al. EMBO J. 17, 4092–4100 (1998).
Altieri A.S., et al. Nature Struct. Biol. 7, 470–474 (2000).
Altieri, A.S. et al. FEBS Lett. 415, 221–226 (1997).
Legault, P., Li, J., Mogridge, J., Kay, L.E. & Greenblatt, J. Cell 93, 289–299 (1998)
Cai, Z. et al. Nature Struct. Biol. 5, 203–212 (1998).
Battiste, J.L. et al. Science 273, 1547–1551 (1996).
Tan, R. & Frankel, A.D. Proc. Natl. Acad. Sci. USA 92, 5282–5287 (1995).
Altschul, S.F., et al. Nucleic Acids Res. 25, 3389–3402 (1997).
Koonin, E.V. Nucleic Acids Res. 22, 2476–2478 (1994)
King, M., Ton, D. & Redman, K.L. Biochem. J. 337, 29–35 (1999).
Reid, R., Greene, P.J. & Santi, D.V. Nucleic Acids Res, 27, 3138–3145 (1999).
Sharrock, R.A., Gourse, R.L. & Nomura, M. Proc. Natl. Acad. Sci. USA 82, 5275–5279 (1985).
McRorie, D.K. & Voelker, P.J. Self-associating systems in the analytical ultracentrifuge (Beckman Instruments Inc., Fullerton, California; 1993).
Gopal, B. et al. Acta Crystallogr D 56, 64–66 (2000).
Otinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1996).
Terwillinger, T.C. & Berendzen, J. Acta Crystallogr. D 55, 849–861 (1999)
Kelywegt, G.J. & Read, R.J. Structure 5, 1557–1569 (1997).
Cowtan, K. Joint CCP4 and ESF-EACBM newsletter on protein crystallography 31, 33–38 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Lamzin, V.S. & Wilson, K.S. Acta Crystallogr. D 49, 129–147 (1993).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Acta Crystallogr. D 53, 240–255 (1997).
Acknowledgements
This work was supported by the Medical Research Council of the UK. We thank S. Gamblin for help in data collection and crystallographic calculations and A. Lane for advice. S. Cole of the Institute Pasteur kindly provided the M. tuberculosis cosmids from which the gene encoding NusB was amplified. This work was supported in part by the European Commission Science Research and Development Programme.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gopal, B., Haire, L., Cox, R. et al. The crystal structure of NusB from Mycobacterium tuberculosis. Nat Struct Mol Biol 7, 475–478 (2000). https://doi.org/10.1038/75876
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/75876
This article is cited by
-
Putative volatile biomarkers of bovine tuberculosis infection in breath, skin and feces of cattle
Molecular and Cellular Biochemistry (2023)
-
Comparative genomics and transcriptional profiles of Saccharopolyspora erythraea NRRL 2338 and a classically improved erythromycin over-producing strain
Microbial Cell Factories (2012)
-
The structure of the transcriptional antiterminator NusB from Escherichia coli
Nature Structural Biology (2000)