Abstract
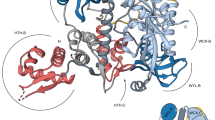
The Escherichia coli Rob protein is a transcription factor belonging to the AraC/XylS protein family that regulates genes involved in resistance to antibiotics, organic solvents and heavy metals. The genes encoding these proteins are activated by the homologous proteins MarA and SoxS, although the level of activation can vary for the different transcription factors. Here we report a 2.7 Å crystal structure of Rob in complex with the micF promoter that reveals an unusual mode of binding to DNA. The Rob–DNA complex differs from the previously reported structure of MarA bound to the mar promoter, in that only one of Rob's dual helix-turn-helix (HTH) motifs engages the major groove of the binding site. Biochemical studies show that sequence specific interactions involving only one of Rob's HTH motifs are sufficient for high affinity binding to DNA. The two different modes of DNA binding seen in crystal structures of Rob and MarA also match the distinctive patterns of DNA protection by AraC at several sites within the pBAD promoter. These and other findings suggest that gene activation by AraC/XylS transcription factors might involve two alternative modes of binding to DNA in different promoter contexts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Skarstad, K., Thony, B., Hwang, D.S. & Kornberg, A. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268, 5365–5370 (1993).
Gallegos, M.T., Schleif, R., Bairoch, A., Hofmann, K. & Ramos, J.L. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410 (1997).
Demple, B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon — a review. Gene 179, 53–57 (1996).
Alekshun, M.N. & Levy, S.B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents. Chemother. 41, 2067–2075 (1997).
Cohen, S.P., Hachler, H. & Levy, S.B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175, 1484–1492 (1993).
Ariza, R.R., Li, Z., Ringstad, N. & Demple, B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177, 1655–1661 (1995).
Nakajima, H., Kobayashi, K., Kobayashi, M., Asako, H. & Aono, R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl. Environ. Microbiol. 61, 2302–2307 (1995).
White, D.G., Goldman, J.D., Demple, B. & Levy, S.B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179, 6122–6126 (1997).
Jair, K.W. et al. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178, 2507–2513 (1996).
Greenberg, J.T., Chou, J.H., Monach, P.A. & Demple, B. Activation of oxidative stress genes by mutations at the soxQ/cfxB/marA locus of Escherichia coli. J. Bacteriol. 173, 4433–4439 (1991).
Ariza, R.R., Cohen, S.P., Bachhawat, N., Levy, S.B. & Demple, B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 176, 143–148 (1994).
Martin, R.G., Gillette, W.K., Rhee, S. & Rosner, J.L. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter [In Process Citation]. Mol. Microbiol. 34, 431–441 (1999).
Rhee, S., Martin, R.G., Rosner, J.L. & Davies, D.R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl Acad. Sci. USA 95, 10413–10418 (1998).
Kakeda, M., Ueguchi, C., Yamada, H. & Mizuno, T. An Escherichia coli curved DNA-binding protein whose expression is affected by the stationary phase-specific sigma factor sigma S. Mol. Gen. Genet. 248, 629–634 (1995).
Lobell, R.B. & Schleif, R.F. DNA looping and unlooping by AraC protein. Science 250, 528–532 (1990).
Ramos, J.L., Michan, C., Rojo, F., Dwyer, D. & Timmis, K. Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J. Mol. Biol. 211, 373–382 (1990).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Thoden, J.B., Ruzicka, F.J., Frey, P.A., Rayment, I. & Holden, H.M. Structural analysis of the H166G site-directed mutant of galactose-1-phosphate uridylyltransferase complexed with either UDP-glucose or UDP-galactose: detailed description of the nucleotide sugar binding site. Biochemistry 36, 1212–1222 (1997).
Li, Z. & Demple, B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 269, 18371–18377 (1994).
Zhang, X., Reeder, T. & Schleif, R. Transcription activation parameters at ara pBAD. J. Mol. Biol. 258, 14–24 (1996).
Jair, K.W., Fawcett, W.P., Fujita, N., Ishihama, A., Wolf, R.E. Jr., Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 19, 307–317 (1996).
Li, Z. & Demple, B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20, 937–945 (1996).
Martin, R.G., Gillette, W.K., Rhee, S. & Rosner, J.L. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Molec. Microbiol. 35, 623–634 (2000).
Niland, P., Huhne, R. & Muller-Hill, B. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 264, 667–674 (1996).
Reeder, T. & Schleif, R. AraC protein can activate transcription from only one position and when pointed in only one direction. J. Mol. Biol. 231, 205–218 (1993).
Brunelle, A. & Schleif, R. Determining residue-base interactions between AraC protein and araI DNA. J. Mol. Biol. 209, 607–622 (1989).
Hendrickson, W. & Schleif, R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl Acad. Sci. USA 82, 3129–3133 (1985).
Martin, R.G., Jair, K.W., Wolf, R.E. Jr. & Rosner, J.L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178, 2216–2223 (1996).
Koh, Y.S., Chung, W.-H., Lee, J.-H. & Roe, J.-H. The reversed SoxS-binding site upstream of the ribA promoter in Escherichia coli. Mol. Gen. Genet. 261, 374–380 (1999).
Nunoshiba, T., Hidalgo, E., Li, Z. & Demple, B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J. Bacteriol. 175, 7492–7494 (1993).
Wood, T.I. et al. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 34, 414–430 (1999).
Doublie, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 461–472 (1997).
Bailey, S. The CCP4 suite: programs for protein crystallography. Acta Crystallog. D 52, 30–42 (1994).
Abrahams, J.P. & Leslie, A.G.W. Methods used in the structure determination of bovine mitochrondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996).
Brunger, A.T. et al. Crystallography,NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A. O: the manual (, Uppsala, Sweden; 1992) http://kaktus.kemi.aau.dk
Brunger, A.T. The free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–474 (1992).
Evans, S.V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11, 134–138, (1993).
Barton, G.J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6, 37–40 (1993).
Acknowledgements
We thank A. Lau, M. Sawaya and members of the Hogle research group for assistance with X-ray data collection. We appreciate the generous gift of purified MarA protein from M. Alekshun and S. Levy (Tufts University School of Medicine). This work was supported by grants from the National Institutes of Health (T.E, B.D.), a Howard Hughes Medical Institute Predoctoral Fellowship (H.J.K.), and the Agrotechnological Research Institute, The Netherlands (M.H.J.B.). We also acknowledge the support of the Giovanni Armenise Foundation for Advanced Scientific Research and the Harvard Center for Structural Biology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kwon, H., Bennik, M., Demple, B. et al. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Mol Biol 7, 424–430 (2000). https://doi.org/10.1038/75213
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/75213
This article is cited by
-
Determining the potential targets of silybin by molecular docking and its antibacterial functions on efflux pumps and porins in uropathogenic E. coli
International Microbiology (2024)
-
Evolutionary engineering of E. coli MG1655 for tolerance against isoprenol
Biotechnology for Biofuels (2020)
-
Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering
Nature Biotechnology (2017)
-
The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity
Nature Communications (2017)
-
GyrI-like proteins catalyze cyclopropanoid hydrolysis to confer cellular protection
Nature Communications (2017)