Abstract
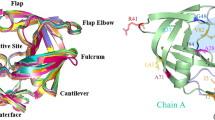
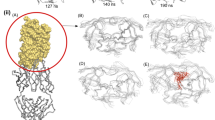
We have used an ‘activated’ molecular dynamics approach to simulate flap opening in HIV-1 protease. An initial impulse for flap opening was provided by applying harmonic constraints to non-flap residues. After an initial ‘melting’ phase, the two β-hairpin structures that constitute the flaps opened to a 25 Å gap within 200 ps of simulation. Analysis of backbone torsion angles suggests that flap opening is related to conformational changes at Lys 45, Met 46, Gly 52 and Phe 53. In contrast, similar molecular dynamics simulations on the M461 mutant, which is associated with drug resistance, indicates that this mutation stabilizes the flaps in a closed conformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Wlodawer, A. & Erickson, J.W. Structure-based inhibitors of HIV-1 protease. A. Rev. Biochem. 62, 543–585 (1993).
Ho, D.D. et al. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J. Virol. 68, 2016–2020 (1994).
Kaplan, A.H. et al. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc .natn .Acad. Sci. U.S.A. 91, 5597–5601 (1994).
Harte, W.E., Jr et al. Domain communication in the dynamical structure of human immunodeficiency virus 1 protease. Proc. natn. Acad. Sci. U.S.A. 87, 8864–8868 (1990).
York, D.M., Darden, T.A., Pedersen, L.G. & Anderson, M.W. Molecular dynamics simulation of HIV-1 protease in a crystalline environment and in solution. Biochemistry 32, 1443–1453 (1993).
Wlodawer, A. et al. Conserved folding in retroviral proteases. Crystal structure of a synthetic HIV-1 protease. Science 245, 616 (1989).
Miller, M. et al. Structure of a complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Å resolution. Science 246, 1149–1152 (1989).
Pearlman, D.A. et al. AMBER 4.0 (San Francisco; 1991).
Wilderspin, A.F. & Sugrue, R.J. Alternative native flap conformation revealed by 2.3 Å resolution structure of SIV proteinase . J. molec. Biol. 239, 97–103 (1994).
Zhao, B. et al. Three-dimensional structure of a simian immunodeficiency virus protease/inhibitor complex. Implications for the design of human immunodeficiency virus type 1 and 2 protease inhibitors. Biochemistry 32, 13054–13060 (1993).
Weiner, S.J., Kollman, P.A., Nguyen, D.T. & Case, D.A. An all atom force field for simulations of proteins and nucleic acids. J. comp. Chem. 7, 230–252 (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Collins, J., Burt, S. & Erickson, J. Flap opening in HIV-1 protease simulated by ‘activated’ molecular dynamics. Nat Struct Mol Biol 2, 334–338 (1995). https://doi.org/10.1038/nsb0495-334
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0495-334
This article is cited by
-
NMR and MD studies combined to elucidate inhibitor and water interactions of HIV-1 protease and their modulations with resistance mutations
Journal of Biomolecular NMR (2019)
-
Effects of HIV-1 protease on cellular functions and their potential applications in antiretroviral therapy
Cell & Bioscience (2012)
-
Molecular dynamics simulation of HIV-protease with polarizable and non-polarizable force fields
Indian Journal of Physics (2009)
-
Structural basis for the resilience of Darunavir (TMC114) resistance major flap mutations of HIV-1 protease
Interdisciplinary Sciences: Computational Life Sciences (2009)
-
Molecular dynamics simulations of 14 HIV protease mutants in complexes with indinavir
Journal of Molecular Modeling (2004)