Abstract
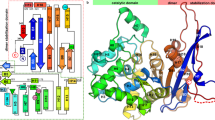
The crystal structure of GMP synthetase serves as a prototype for two families of metabolic enzymes. The Class I glutamine amidotransferase domain of GMP synthetase is found in related enzymes of the purine, pyrimidine, tryptophan, arginine, histidine and folic acid biosynthetic pathways. This domain includes a conserved Cys-His-Glu triad and is representative of a new family of enzymes that use a catalytic triad for enzymatic hydrolysis. The structure and conserved sequence fingerprint of the nucleotide-binding site in a second domain of GMP synthetase are common to a family of ATP pyrophosphatases, including NAD synthetase, asparagine synthetase and argininosuccinate synthetase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Hyde, C.C., Ahmed, S.A., Padlan, E.A., Miles, E.W. & Davies, D.R. Three-dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium . J. biol. Chem. 263, 17857–17871 (1988).
Wilson, K.P., Shewchuk, L.M., Brennan, R.G., Otsuka, A.J. & Matthews, B.W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. natn. Acad. Sci. U.S.A. 89, 9257–9261 (1992).
Zalkin, H. The amidotransferases. Adv. Enzymol. Relat Areas molec. Biol. 66, 203–309 (1993).
Fukuyama, T.T. & Moyed, H.S. A separate antibiotic binding site in xanthosine-5'-phosphate aminase: Inhibitor- and substrate-binding studies. Biochemistry 3, 1488–1492 (1964).
von der Saal, W., Crysler, C.S. & Villafranca, J.J. Positional isotope exchange and kinetic experiments with Escherichia coli guanosine 5'-monophosphate synthetase. Biochemistry 24, 5343–5350 (1985).
Lusty, C.J. Detection of an enzyme bound γ-glutamyl acyl ester of carbamyl phosphate synthetase of Escherichia coli . FEBS Lett. 314, 135–138 (1992).
Roux, B. & Walsh, C.T. p-Aminobenzoate synthesis in Escherichia coli: kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry 31, 6904–6910 (1992).
Amuro, N., Paluh, J.L. & Zalkin, H. Replacement by site-directed mutagenesis indicates a role for histidine 170 in the glutamine amide transfer function of anthranilate synthase. J. biol. Chem. 260, 14844–14849 (1985).
Willison, J.C. Biochemical genetics revisited: the use of mutants to study carbon and nitrogen metabolism in the photosynthetic bacteria. FEMS microbiol. Rev. 104, 1–38 (1993).
Bork, P. & Koonin, E.V. A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins 20, 347–355 (1994).
Van Lookeren Campagne, M.M., Franke, J. & Kessin, R.H. Functional cloning of a Dictyostelium discoideum cDNA encoding GMP synthetase. J. biol. Chem. 266, 16448–16452 (1991).
Hirst, M., Haliday, E., Nakamura, J. Lou, L. Human GMP synthetase. Protein purification, cloning, and functional expression of cDNA. J. biol. Chem. 269, 23830–23837 (1994).
Sakamoto, N., Hatfield, G.W. & Moyed, H.S. Physical properties and subunit structure of xanthosine 5'-phosphate aminase. J. biol. Chem. 247, 5880–5887 (1972).
Tesmer, J.J.G., Stemmler, T.L., Penner-Hahn, J.E., Davisson, V.J. & Smith, J.L. Preliminary X-ray analysis of Escherichia coli GMP synthetase: determination of anomalous scattering factors for a cysteinyl mercury derivative. Proteins 18, 394–403 (1994).
Oillis, D.L. et al. The α/β hydrolase fold. Prot. Engng. 5, 197–211 (1992).
Yee, V.C. et al. Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII. Proc. natn. Acad. Sci. U.S.A. 91, 7296–7300 (1994).
Wei, Y. et al. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nature struct. Biol. 2, 218–223 (1995).
Roux, B. & Walsh, C.T. p-Aminobenzoate synthesis in Escherichia coli: mutational analysis of three conserved amino acid residues of the amidotransferase PabA. Biochemistry 32, 3763–3768 (1993).
Sprang, S. et al. The three-dimensional structure of asn102 mutant of trypsin: role of asp102 in serine protease catalysis. Science 237, 905–909 (1987).
Carter, P. & Wells, J.A. Dissecting the catalytic triad of a serine protease. Nature 332, 564–568 (1988).
McGrath, M.E., Wilke, M.E., Higaki, J.N., Craik, C.S. & Fletterick, R.J. Crystal structures of two engineered thiol trypsins. Biochemistry 28, 9264–9270 (1989).
Corey, D.R. & Craik, C.S. An investigation into the minimum requirements for peptide hydrolysis by mutation of the catalytic triad of trypsin. J. Am. chem. Soc 114, 1784–1790 (1992).
Matthews, D.A. et al. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77, 761–771 (1994).
Rossmann, M.G., Liljas, A., Bränden, C.-I. & Banaszak, L.J. in The Enzymes (ed. Boyer, P.D.) 61–102 (Academic Press, New York, 1975).
Schulz, G.E. Binding of nucleotides by proteins. Curr. Opin. struct. Biol. 2, 61–67 (1992).
Sturrock, S.S. & Collins, J.F. MPsrch version 1.5 (University of Edinburgh, UK, 1993).
Perona, J., Rould, M.A. & Steitz, T.A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry 32, 8758–8771 (1993).
Kobayashi, K., Jackson, M.J., Tick, D.B., O'Brien, W.E. & Beaudet, A.L. Heterogeneity of mutations in argininosuccinate synthetase causing human citrullinemia. J. biol. Chem. 265, 11361–11367 (1990).
Kobayashi, K., Rosenbloom, C., Beaudet, A.L. & O'Brien, W.E. Additional mutations in argininosuccinate synthetase causing citrullinemia. Mol. biol.Med. 8, 95–100 (1991).
Scapin, G., Grubmeyer, C & Sacchettini, J.C. Crystal structure of orotate phosphoribosyltransferase. Biochemistry 33, 1288–1294 (1994).
Smith, J.L. et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science 264, 1427–1433 (1994).
Eads, J.C., Scapin, G., Xu, Y., Grubmeyer, C. & Sacchettini, J.C. The crystal structure of human hypoxanthine-guanine phosphoribosyltransferase. Cell 78, 325–334 (1994).
Spencer, R.L. & Preiss, J. Biosynthesis of diphosphopyridine nucleotide: The purification and the properties of diphosphopyridine nucleotide synthetase from Escherichia coli . J. biol. Chem. 242, 385–392 (1967).
Suyama, M., Ogiwara, A., Nishioka, T. & Oda, J. Searching for amino acid motifs among enzymes: the enzyme-reaction database. Comput. appl. Biosci. 9, 9–15 (1993).
Hirai, K., Matsuda, Y. & Nakagawa, H. Purification and characterization of GMP synthetase from Yoshida sarcoma ascites cells. J. Biochem. 102, 893–902 (1987).
Nakamura, J. & Lou, L. Biochemical characterization of human GMP synthetase. J. biol. Chem. 270, 7347–7353 (1995).
Zalkin, H. & Truitt, C.D. Characterization of the glutamine site of Escherichia coli guanosine 5'-monophosphate synthetase. J. biol. Chem. 252, 5431–5436 (1977).
Patel, N., Moyed, H.S. & Kane, J.F. Properties of xanthosine 5'-monophosphate-amidotransferase from Escherichia coli . Arch. Biochem. Biophys. 178, 652–661 (1977).
Zyk, N., Citri, N. & Moyed, H.S. Conformative response of xanthosine 5'-phosphate aminase. Biochemistry 8, 2787–2794 (1969).
Anderson, C.M., Zucker, F.H. & Steitz, T.A. Space-filling models of kinase clefts and conformation changes. Science 204, 375–380 (1979).
Liao, D.-I., Karpusas, M. & Remington, S.J. Crystal structure of an open conformation of citrate synthase from chicken heart at 2. 8-Å resolution. Biochemistry 30, 6031–6036 (1991).
Vonrhein, C., Schlauderer, G.J. & Schulz, G.E. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinase. Structure 3, 483–490 (1995).
Hendrickson, W.A., Horton, J.R. & LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990).
Nair, V. & David, A.J. A new synthesis of isoguanosine. J. org. Chem. 50, 406–408 (1985).
Mishra, N.C. & Broom, A.D. A novel synthesis of nucleoside 5'-triphosphates. J. Chem. Soc chem. Commun. 1991, 1276 (1991).
Hamlin, R. Multiwire area X-ray diffractometers. Meth. Enzymol. 114, 416–452 (1985).
Howard, A.J., Nielsen, C. & Xuong, N.H. Software for a diffractometer with multiwire area detector. Meth. Enzymol. 114, 452–472 (1985).
Kabsch, W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. appl. Crystallogr. 21, 916–924 (1988).
Bailey, S. The CCP4 suite-programs for protein crystallography. Acta crystallogr. D50, 760–763 (1994).
Sakabe, N. X-ray diffraction data collection system for modern protein crystallography with a Weissenberg camera and an imaging plate using synchrotron radiation. Nucl. Instr. Meth. A303, 448–463 (1991).
Otwinowski, Z. in Data Collection and Processing (ed. Sawyer, N.I. & Bailey, S.) 56–62 (Science and Engineering Research Council Daresbury Laboratory, Daresbury, U.K., 1993).
Quiocho, F.A. & Richards, F.M. Intermolecular cross linking of a protein in the crystalline state: carboxypeptidase-A. Proc. natn. Acad. Sci. U.S.A. 52, 833 (1964).
Ringe, D., Petsko, G.A., Yamakura, F., Suzuki, K. & Ohmori, D. Structure of iron superoxide dismutase from Pseudomonas ovalis at 2. 9-Å resolution. Proc. natn. Acad. Sci. U.S.A. 80, 3879–3883 (1983).
Otwinowski, Z. in Isomorphous Replacement and Anomalous Scattering (ed. Wolf, W. & Leslie, A.G.W.) 80–86 (Science and Engineering Research Council Daresbury Laboratory, Daresbury, U. K., 1991).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta crystallogr. A47, 110–119 (1991).
Brünger, A.T. X-PLOR Version 3.1 A system for X-ray Crystallography and NMR 1–382 (Yale University Press, New Haven and London, 1992).
Brünger, A.T., Krukowski, A. & Erickson, J.W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta crystallogr. A46, 585–593 (1990).
Engh, R.A. & Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta crystallogr. A47, 392–400 (1991).
Tiedeman, A.A., Smith, J.M. & Zalkin, H. Nucleotide sequence of the guaA gene encoding GMP synthetase of Escherichia coli K12. J. biol. Chem. 260, 8676–8679 (1985).
Mäntsälä, P. & Zalkin, H. Cloning and sequence of Bacillus subtilis pur A and guaA, involved in the conversion of IMP to AMP and GMP. J. Bacteriol. 174, 1883–1890 (1992).
Dujardin, G., Kermorgant, M., Slonimski, P.P. & Boucherie, H. Cloning and sequencing of the GMP synthetase-encoding gene of Saccharomycescerevisiae . Gene 139, 127–132 (1994).
Smith, J.L. Structures of glutamine amidotransferases from the purine biosynthetic pathway. Biochem. Soc. Trans. 23, 894–898 (1995).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Schroeder, E., Phillips, C., Garman, E., Harlos, K. & Crawford, C. X-ray crystallographic structure of a papain leupeptin complex. FEBS Lett. 315, 38–42 (1993).
Baker, E.N. & Drenth, J. in Biological Macromolecules and Assemblies, Vol. 3 (eds. Jurnak, F.A. & McPherson, A.) 314–368 (Wiley, New York, 1987).
Marquart, M., Walter, J., Deisenhofer, J., Bode, W. & Huber, R. The geometry of the reactive site and of the peptide groups in trypsin, trypsinogen and its complexes with inhibitors. Acta crystallogr. b39, 480–490 (1983).
Bullock, T.L., Branchaud, B. & Remington, S.J. Structure of the complex of L-benzylsuccinate with wheat serine carboxypeptidase II at 2. 0-Å resolution. Biochemistry 33, 11127–11134 (1994).
Nishikawa, K. & Ooi, T. Comparison of homologous tertiary structures of proteins. J. theor. Biol. 43, 351–374 (1974).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tesmer, J., Klem, T., Deras, M. et al. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat Struct Mol Biol 3, 74–86 (1996). https://doi.org/10.1038/nsb0196-74
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb0196-74
This article is cited by
-
GATD3A, a mitochondrial deglycase with evolutionary origins from gammaproteobacteria, restricts the formation of advanced glycation end products
BMC Biology (2022)
-
The pathogenic mechanism of Mycobacterium tuberculosis: implication for new drug development
Molecular Biomedicine (2022)
-
Molecular basis for the allosteric activation mechanism of the heterodimeric imidazole glycerol phosphate synthase complex
Nature Communications (2021)
-
Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation
Nature Communications (2020)
-
High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity
Communications Biology (2019)