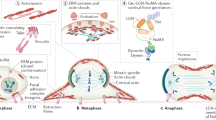
Key Points
-
Cleavage plane angle is a developmentally regulated property in mammalian neural progenitor cells. It is highly correlated with the production of different types of daughter cells and can be used to control the distribution of cell fate determinants.
-
Tumor suppressor proteins help regulate the asymmetric distribution of various proteins necessary for ganglion mother cell fate specification in the dividing Drosophila melanogaster neuroblast.
-
Asymmetrical inheritance of vertebrate numb homologues during neural progenitor cell division has important consequences for cell fate determination.
-
A conserved cassette of apically localized proteins helps to establish distinct apical and basal domains in D. melanogaster neuroblasts. The proteins also help to coordinate alignment of the mitotic spindle between these domains.
-
D. melanogaster partner of inscuteable (PINS) links spindle orientation to the apical protein domain and also mediates the establishment of spindle asymmetry. Mammalian homologs of PINS are involved in regulating spindle orientation with important implications for daughter cell fate.
-
The heterotrimeric G proteins transduce signals necessary for the control of spindle alignment in flies and mammals. Their cycle of activity is closely controlled by a number of regulatory proteins.
-
D. melanogaster mushroom body defect (MUD) and a mammalian protein that shares limited homology, nuclear mitotic apparatus (NUMA), serve as links between cortical proteins and the mitotic spindle, allowing them to coordinate.
-
The presence of adherens junctions in D. melanogaster neuroepithelial cells influences spindle orientation. Several animal models indicate that adherens junctions and associated polarity proteins might have important roles in determining division mode and the identity of mammalian neural progenitors.
Abstract
The mitotic spindle is the cellular scaffold that facilitates proper segregation of genetic material during cell division. Far from being static, the spindle is a dynamically regulated tool that can alter its size, shape and position during mitosis. Work in both insect and vertebrate systems has shown that regulation of this structure involves an array of highly conserved proteins. Moreover, it is now clear that tight regulation of the spindle during the process of neurogenesis is paramount to proper cell division and generation of the nervous system as a whole.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gotz, M. & Huttner, W. B. The cell biology of neurogenesis. Nature Rev. Mol. Cell Biol. 6, 777–788 (2005).
Takahashi, T., Nowakowski, R. S. & Caviness, V. S. The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J. Neurosci. 16, 6183–6196 (1996).
Caviness, V. S., Takahashi, T. & Nowakowski, R. S. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 18, 379–383 (1995).
Langman, J., Guerrant, R. L. & Freeman, B. G. Behavior of neuro-epithelial cells during closure of the neural tube. J. Comp. Neurol. 127, 399–411 (1966).
Martin, A. H. Significance of mitotic spindle fibre orientation in the neural tube. Nature 216, 1133–1134 (1967).
Chenn, A. & McConnell, S. K. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell 82, 631–641 (1995). Through live imaging of cultured brain slices, this paper provides the first evidence that division orientation of progenitor cells in the mammalian VZ is regulated in a temporal manner.
Haydar, T. F., Ang, E. & Rakic, P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc. Natl Acad. Sci. USA 100, 2890–2895 (2003).
Cayouette, M., Whitmore, A. V., Jeffery, G. & Raff, M. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J. Neurosci. 21, 5643–5651 (2001).
Cayouette, M. & Raff, M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development 130, 2329–2339 (2003).
Adams, R. J. Metaphase spindles rotate in the neuroepithelium of rat cerebral cortex. J. Neurosci. 16, 7610–7618 (1996).
Reid, C. B., Tavazoie, S. F. & Walsh, C. A. Clonal dispersion and evidence for asymmetric cell division in ferret cortex. Development 124, 2441–2450 (1997).
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720 (2001).
Silva, A. O., Ercole, C. E. & McLoon, S. C. Plane of cell cleavage and numb distribution during cell division relative to cell differentiation in the developing retina. J. Neurosci. 22, 7518–7525 (2002).
Smart, I. H. Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: a pilot study based on recording the number, location and plane of cleavage of mitotic figures. J. Anat. 116, 67–91 (1973).
Zamenhof, S. Quantitative studies of mitoses in fetal rat brain: orientations of the spindles. Brain Res. 428, 143–146 (1987).
Das, T., Payer, B., Cayouette, M. & Harris, W. A. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron 37, 597–609 (2003).
Wodarz, A. & Huttner, W. B. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech. Dev. 120, 1297–1309 (2003).
Nelson, W. J. Adaptation of core mechanisms to generate cell polarity. Nature 422, 766–774 (2003).
Huttner, W. B. & Kosodo, Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 17, 648–657 (2005).
Huttner, W. B. & Brand, M. Asymmetric division and polarity of neuroepithelial cells. Curr. Opin. Neurobiol. 7, 29–39 (1997).
Haubensak, W., Attardo, A., Denk, W. & Huttner, W. B. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl Acad. Sci. USA 101, 3196–3201 (2004).
Kosodo, Y. et al. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. Embo J. 23, 2314–2324 (2004). Describes a correlation between asymmetrical distribution of apical membrane components with asymmetric divisions of mouse progenitor cells in the embryonic VZ.
Kaltschmidt, J. A., Davidson, C. M., Brown, N. H. & Brand, A. H. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nature Cell Biol. 2, 7–12 (2000).
Spana, E. P. & Doe, C. Q. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121, 3187–3195 (1995).
Hirata, J., Nakagoshi, H., Nabeshima, Y. & Matsuzaki, F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377, 627–630 (1995).
Knoblich, J. A., Jan, L. Y. & Jan, Y. N. Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624–627 (1995).
Vaessin, H. et al. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, 941–953 (1991).
Doe, C. Q., Chu-LaGraff, Q., Wright, D. M. & Scott, M. P. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464 (1991).
Li, L. & Vaessin, H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 14, 147–151 (2000).
Liu, T. H., Li, L. & Vaessin, H. Transcription of the Drosophila CKI gene dacapo is regulated by a modular array of cis-regulatory sequences. Mech. Dev. 112, 25–36 (2002).
Ikeshima-Kataoka, H., Skeath, J. B., Nabeshima, Y., Doe, C. Q. & Matsuzaki, F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–629 (1997).
Shen, C. P., Jan, L. Y. & Jan, Y. N. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90, 449–458 (1997).
Betschinger, J., Mechtler, K. & Knoblich, J. A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 (2006).
Bello, B., Reichert, H. & Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648 (2006).
Lee, C. Y., Wilkinson, B. D., Siegrist, S. E., Wharton, R. P. & Doe, C. Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 (2006).
Ohshiro, T., Yagami, T., Zhang, C. & Matsuzaki, F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408, 593–596 (2000).
Peng, C. Y., Manning, L., Albertson, R. & Doe, C. Q. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408, 596–600 (2000).
Albertson, R. & Doe, C. Q. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nature Cell Biol. 5, 166–170 (2003).
Uemura, T., Shepherd, S., Ackerman, L., Jan, L. Y. & Jan, Y. N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 (1989). Describes the original cloning of numb, showing its relevance to guiding cell fate decisions in the D. melanogaster nervous system.
Rhyu, M. S., Jan, L. Y. & Jan, Y. N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994).
Spana, E. P., Kopczynski, C., Goodman, C. S. & Doe, C. Q. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development 121, 3489–3494 (1995).
Spana, E. P. & Doe, C. Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996).
Guo, M., Jan, L. Y. & Jan, Y. N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 (1996).
Frise, E., Knoblich, J. A., Younger-Shepherd, S., Jan, L. Y. & Jan, Y. N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell–cell interaction in sensory organ lineage. Proc. Natl Acad. Sci. USA 93, 11925–11932 (1996).
Roegiers, F. & Jan, Y. N. Asymmetric cell division. Curr. Opin. Cell Biol. 16, 195–205 (2004).
Betschinger, J. & Knoblich, J. A. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14, R674–R685 (2004).
Zhong, W., Feder, J. N., Jiang, M. M., Jan, L. Y. & Jan, Y. N. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53 (1996). Shows that mNUMB can functionally substitute for numb and that its inheritance in dividing progenitors is dependent on spindle orientation.
Zhong, W. et al. Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl Acad. Sci. USA 97, 6844–6849 (2000).
Wakamatsu, Y., Maynard, T. M., Jones, S. U. & Weston, J. A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71–81 (1999).
Petersen, P. H., Zou, K., Hwang, J. K., Jan, Y. N. & Zhong, W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 (2002).
Petersen, P. H., Zou, K., Krauss, S. & Zhong, W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nature Neurosci. 7, 803–811 (2004).
Li, H. S. et al. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 40, 1105–1118 (2003).
Shen, Q., Zhong, W., Jan, Y. N. & Temple, S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853 (2002).
Castaneda-Castellanos, D. R. & Kriegstein, A. R. Controlling neuron number: does Numb do the math? Nature Neurosci. 7, 793–794 (2004).
Austin, C. P., Feldman, D. E., Ida, J. A. & Cepko, C. L. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development 121, 3637–3650 (1995).
Henrique, D. et al. Maintenance of neuroepithelial progenitor cells by Delta–Notch signalling in the embryonic chick retina. Curr. Biol. 7, 661–670 (1997).
Yu, F. et al. A mouse homologue of Drosophila pins can asymmetrically localize and substitute for pins function in Drosophila neuroblasts. J. Cell Sci. 116, 887–896 (2003).
Wodarz, A., Ramrath, A., Kuchinke, U. & Knust, E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544–547 (1999).
Schober, M., Schaefer, M. & Knoblich, J. A. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402, 548–551 (1999).
Yu, F., Morin, X., Cai, Y., Yang, X. & Chia, W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 (2000).
Petronczki, M. & Knoblich, J. A. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nature Cell Biol. 3, 43–49 (2001).
Wodarz, A., Ramrath, A., Grimm, A. & Knust, E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150, 1361–1374 (2000).
Rolls, M. M., Albertson, R., Shih, H. P., Lee, C. Y. & Doe, C. Q. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, 1089–1098 (2003).
Shen, C. P. et al. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 12, 1837–1846 (1998).
Li, P., Yang, X., Wasser, M., Cai, Y. & Chia, W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 90, 437–447 (1997).
Kuchinke, U., Grawe, F. & Knust, E. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 8, 1357–1365 (1998).
Kraut, R. & Campos-Ortega, J. A. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev. Biol. 174, 65–81 (1996).
Kraut, R., Chia, W., Jan, L. Y., Jan, Y. N. & Knoblich, J. A. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50–55 (1996).
Schaefer, M., Shevchenko, A. & Knoblich, J. A. A protein complex containing Inscuteable and the Gα-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353–362 (2000).
Parmentier, M. L. et al. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20, RC84 (2000).
Lee, C. Y., Robinson, K. J. & Doe, C. Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594–598 (2006).
Betschinger, J., Mechtler, K. & Knoblich, J. A. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326–330 (2003).
Cai, Y., Yu, F., Lin, S., Chia, W. & Yang, X. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51–62 (2003).
Blumer, J. B., Chandler, L. J. & Lanier, S. M. Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. Localization of LGN to the midbody during cytokinesis. J. Biol. Chem. 277, 15897–15903 (2002).
Sanada, K. & Tsai, L. H. G protein βγ subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119–131 (2005). Demonstrates a conserved role for non-canonical G-protein signalling in orienting spindles of mammalian progenitors and shows that spindle orientation has a role in determining daughter cell fate.
Zigman, M. et al. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron 48, 539–545 (2005).
Lechler, T. & Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280 (2005).
Siderovski, D. P. & Willard, F. S. The GAPs, GEFs, and GDIs of heterotrimeric G-protein α subunits. Int. J. Biol. Sci. 1, 51–66 (2005).
Schaefer, M., Petronczki, M., Dorner, D., Forte, M. & Knoblich, J. A. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194 (2001).
Yu, F., Cai, Y., Kaushik, R., Yang, X. & Chia, W. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 162, 623–633 (2003).
Yu, F. et al. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes Dev. 19, 1341–1353 (2005).
Fuse, N., Hisata, K., Katzen, A. L. & Matsuzaki, F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr. Biol. 13, 947–954 (2003).
Izumi, Y., Ohta, N., Itoh-Furuya, A., Fuse, N. & Matsuzaki, F. Differential functions of G protein and Baz–aPKC signaling pathways in Drosophila neuroblast asymmetric division. J. Cell Biol. 164, 729–738 (2004).
Wang, H. et al. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nature Cell Biol. 7, 1091–1098 (2005).
Hampoelz, B., Hoeller, O., Bowman, S. K., Dunican, D. & Knoblich, J. A. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nature Cell Biol. 7, 1099–1105 (2005). Along with reference 84, this paper describes a role for the GEF RIC8 in regulating neuroblast spindle asymmetry and G-protein subunit localization and activity.
Kaushik, R., Yu, F., Chia, W., Yang, X. & Bahri, S. Subcellular localization of LGN during mitosis: evidence for its cortical localization in mitotic cell culture systems and its requirement for normal cell cycle progression. Mol. Biol. Cell 14, 3144–3155 (2003).
Cismowski, M. J., Takesono, A., Bernard, M. L., Duzic, E. & Lanier, S. M. Receptor-independent activators of heterotrimeric G-proteins. Life Sci. 68, 2301–2308 (2001).
Takesono, A. et al. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J. Biol. Chem. 274, 33202–33205 (1999).
Natochin, M. et al. AGS3 inhibits GDP dissociation from Gα subunits of the Gi family and rhodopsin-dependent activation of transducin. J. Biol. Chem. 275, 40981–40985 (2000).
Peterson, Y. K. et al. Stabilization of the GDP-bound conformation of Giα by a peptide derived from the G-protein regulatory motif of AGS3. J. Biol. Chem. 275, 33193–33196 (2000).
De Vries, L. et al. Activator of G protein signaling 3 is a guanine dissociation inhibitor for Gαi subunits. Proc. Natl Acad. Sci. USA 97, 14364–14369 (2000).
Bernard, M. L., Peterson, Y. K., Chung, P., Jourdan, J. & Lanier, S. M. Selective interaction of AGS3 with G-proteins and the influence of AGS3 on the activation state of G-proteins. J. Biol. Chem. 276, 1585–1593 (2001).
Tonissoo, T., Meier, R., Talts, K., Plaas, M. & Karis, A. Expression of ric-8 (synembryn) gene in the nervous system of developing and adult mouse. Gene Expr. Patterns 3, 591–594 (2003).
Tall, G. G., Krumins, A. M. & Gilman, A. G. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 278, 8356–8362 (2003).
Tall, G. G. & Gilman, A. G. Resistance to inhibitors of cholinesterase 8A catalyzes release of Gαi-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Gαi-GDP complexes. Proc. Natl Acad. Sci. USA 102, 16584–16589 (2005).
Shinohara, H. et al. Gi2 signaling enhances proliferation of neural progenitor cells in the developing brain. J. Biol. Chem. 279, 41141–41148 (2004).
Prokop, A. & Technau, G. M. Normal function of the mushroom body defect gene of Drosophila is required for the regulation of the number and proliferation of neuroblasts. Dev. Biol. 161, 321–337 (1994).
Siller, K. H., Cabernard, C. & Doe, C. Q. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nature Cell Biol. 8, 594–600 (2006).
Bowman, S. K., Neumuller, R. A., Novatchkova, M., Du, Q. & Knoblich, J. A. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 (2006).
Izumi, Y., Ohta, N., Hisata, K., Raabe, T. & Matsuzaki, F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nature Cell Biol. 8, 586–593 (2006). Along with references 98 and 99, this paper describes the interaction of MUD with the PINS–Gα complex and the role of this complex in coordinating the orientation of mitotic spindles with cortical domain cues.
Zeng, C. NuMA: a nuclear protein involved in mitotic centrosome function. Microsc. Res. Tech. 49, 467–477 (2000).
Du, Q., Stukenberg, P. T. & Macara, I. G. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nature Cell Biol. 3, 1069–1075 (2001).
Du, Q., Taylor, L., Compton, D. A. & Macara, I. G. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 12, 1928–1933 (2002).
Du, Q. & Macara, I. G. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 (2004).
Lu, B., Roegiers, F., Jan, L. Y. & Jan, Y. N. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature 409, 522–525 (2001). Demonstrates that adherens junctions provide a planar spindle orientation cue in Drosophila neuroepithelial cells that, when lost, allows for the randomization of spindle orientation.
Wang, F., Dumstrei, K., Haag, T. & Hartenstein, V. The role of DE-cadherin during cellularization, germ layer formation and early neurogenesis in the Drosophila embryo. Dev. Biol. 270, 350–363 (2004).
Yamashita, Y. M., Jones, D. L. & Fuller, M. T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 (2003).
Le Borgne, R., Bellaiche, Y. & Schweisguth, F. Drosophila E-cadherin regulates the orientation of asymmetric cell division in the sensory organ lineage. Curr. Biol. 12, 95–104 (2002).
Harris, T. J. & Peifer, M. Adherens junction-dependent and-independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167, 135–147 (2004).
Hutterer, A., Betschinger, J., Petronczki, M. & Knoblich, J. A. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell 6, 845–854 (2004).
Muller, H. A. & Wieschaus, E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134, 149–163 (1996).
Harris, T. J. & Peifer, M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170, 813–823 (2005).
Noctor, S. C., Martinez-Cerdeno, V., Ivic, L. & Kriegstein, A. R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neurosci. 7, 136–144 (2004).
Calegari, F., Haubensak, W., Haffner, C. & Huttner, W. B. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 25, 6533–6538 (2005).
Chenn, A., Zhang, Y. A., Chang, B. T. & McConnell, S. K. Intrinsic polarity of mammalian neuroepithelial cells. Mol. Cell Neurosci. 11, 183–193 (1998).
Shoukimas, G. M. & Hinds, J. W. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J. Comp. Neurol. 179, 795–830 (1978).
Hinds, J. W. & Ruffett, T. L. Cell proliferation in the neural tube: an electron microscopic and golgi analysis in the mouse cerebral vesicle. Z. Zellforsch. Mikrosk. Anat. 115, 226–264 (1971).
Nagele, R. G. & Lee, H. Y. Ultrastructural changes in cells associated with interkinetic nuclear migration in the developing chick neuroepithelium. J. Exp. Zool. 210, 89–106 (1979).
Miyata, T. et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131, 3133–3145 (2004).
Chenn, A. & Walsh, C. A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002).
Lien, W. H., Klezovitch, O., Fernandez, T. E., Delrow, J. & Vasioukhin, V. αE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science 311, 1609–1612 (2006). Along with reference 120, this paper shows a crucial role for adherens junction signalling in the regulation of cell proliferation and neurogenesis in the embryonic mammalian brain.
Joberty, G., Petersen, C., Gao, L. & Macara, I. G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nature Cell Biol. 2, 531–539 (2000).
Lin, D. et al. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nature Cell Biol. 2, 540–547 (2000).
Noda, Y. et al. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 6, 107–119 (2001).
Aaku-Saraste, E., Hellwig, A. & Huttner, W. B. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure — remodeling of the neuroepithelium prior to neurogenesis. Dev. Biol. 180, 664–679 (1996).
Manabe, N. et al. Association of ASIP/mPAR-3 with adherens junctions of mouse neuroepithelial cells. Dev. Dyn. 225, 61–69 (2002).
Cappello, S. et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nature Neurosci. 9, 1099–1107 (2006).
Afonso, C. & Henrique, D. PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. J. Cell Sci. 119, 4293–4304 (2006).
Imai, F. et al. Inactivation of aPKCλ results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development 133, 1735–1744 (2006).
Plant, P. J. et al. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nature Cell Biol. 5, 301–308 (2003).
Yamanaka, T. et al. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13, 734–743 (2003).
Yasumi, M. et al. Direct binding of Lgl2 to LGN during mitosis and its requirement for normal cell division. J. Biol. Chem. 280, 6761–6765 (2005).
Klezovitch, O., Fernandez, T. E., Tapscott, S. J. & Vasioukhin, V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 18, 559–571 (2004).
Siegrist, S. E. & Doe, C. Q. Microtubule-induced Pins/Gαi cortical polarity in Drosophila neuroblasts. Cell 123, 1323–1335 (2005). Uncovers a microtubule-dependent pathway that coordinates the alignment of mitotic spindles with PINS–Gα crescents through a DLG/kinesin heavy chain 73-mediated pathway.
Siegrist, S. E. & Doe, C. Q. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133, 529–536 (2006).
Acknowledgements
We would like to thank the following individuals for their comments and input during the writing of this manuscript: Z. Xie, R. Ayala, X. Ge and K. Sanada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Glossary
- Ventricular zone
-
(VZ). The proliferative region of the mammalian neocortex from which neurons arise. It is the most apical layer of the cortex. It lines the cerebral ventricles and sits directly below the subventricular zone.
- Neuroepithelial cell
-
The main proliferative cell type of the early neocortex, which divides to expand the ventricular zone and gives rise to neurons and radial glial cells. Dividing cells of the retina and neural tube are also referred to as neuroepithelial cells.
- Radial glia
-
The main neurogenic cell type found in the neocortical ventricular zone during the peak period of neurogenesis. These cells can divide asymmetrically to generate a neuron as well as another radial glial cell.
- Basal progenitors
-
A class of progenitors which divide at positions basal to the ventricular surface. They usually divide only 1–2 times to produce terminally differentiated neurons. Unlike other neural progenitors, they do not maintain contact with the ventricular surface and lack overt polarity.
- Cleavage plane
-
Refers to the orientation of the cleavage furrow generated during mitosis. The cleavage plane angle is perpendicular to the orientation of the spindle poles and approximately perpendicular to the plane across which two cells separate.
- Horizontal cleavage
-
A mitotic division during which the mitotic spindle poles are oriented along the apicobasal axis, perpendicular to the luminal surface so that the cleavage furrow forms parallel to the luminal surface.
- Vertical cleavage
-
A mitotic division during which the mitotic spindle poles are oriented parallel to the plane of the luminal surface so that the cleavage furrow forms perpendicular to the luminal surface.
- Symmetric division
-
A mitotic division generating daughter cells with identical cell fates.
- Asymmetric division
-
A mitotic division generating daughter cells with different cell fates.
- Neurogenic division
-
Any division that produces a neuron. Terminal neurogenic divisions produce two neurons incapable of further division.
- Tumour suppressor protein
-
A protein that functions to prevent tumour formation by any of a number of mechanisms, including preventing cell cycle re-entry or inducing apoptosis in cells harbouring DNA damage.
- G-protein coupled receptor
-
(GPCR). Any of a family of seven transmembrane proteins that couple extracellular ligand binding to activation of intracellular G-protein signalling cascades. These proteins are involved in signalling processes as diverse as sensory perception, the inflammatory response and autonomic nervous system responses.
- Guanine dissociation inhibitor
-
(GDI). A protein that acts to slow the spontaneous exchange of GDP for GTP on Gα proteins.
- Guanine nucleotide exchange factor
-
(GEF). A protein that catalyses the dissociation of GDP from small Gα proteins to promote binding of GTP.
- Regulator of G-protein signalling
-
(RGS). Any member of a family of proteins that contains an RGS-box domain and can accelerate the intrinsic GTP hydrolysis activity of small Gα proteins.
- GTPase activating proteins
-
(GAP). Proteins, such as regulators of G-protein signalling, that can accelerate the intrinsic GTP hydrolysis activity of small Gα proteins.
- Mushroom body
-
A paired neuropil structure found in the Drosophila melanogaster brain that functions in learning and memory.
- Cadherins
-
A family of adherens junction component proteins that form calcium-dependent homotypic associations across cells to promote cell–cell adhesion and intercellular signalling.
- αE-catenin
-
A member of the α-catenin family expressed in neural progenitor cells. α-Catenins are actin-binding proteins associated with adherens junctions.
Rights and permissions
About this article
Cite this article
Buchman, J., Tsai, LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci 8, 89–100 (2007). https://doi.org/10.1038/nrn2058
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn2058
This article is cited by
-
18q22.1-qter deletion and 4p16.3 microduplication in a boy with speech delay and mental retardation: case report and review of the literature
Molecular Cytogenetics (2018)
-
ASPM regulates symmetric stem cell division by tuning Cyclin E ubiquitination
Nature Communications (2015)
-
Cortical neurogenesis in the absence of centrioles
Nature Neuroscience (2014)
-
TPX2: of spindle assembly, DNA damage response, and cancer
Cellular and Molecular Life Sciences (2014)
-
MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1–Cdc25 pathway
Nature Cell Biology (2011)