Key Points
-
14-3-3 proteins are involved in the control of the cell cycle, transcription and apoptosis. Owing to their multiple interactions with various kinases, receptors, enzymes and structural and cytoskeletal proteins. Although the precise role of 14-3-3 proteins is not fully understood, they seem to control the subcellular localization of proteins and to function as adaptor molecules, stimulating protein–protein interactions.
-
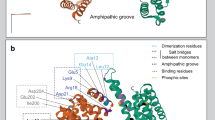
There are seven known members of the 14-3-3 family, but genomic analysis points to the existence of several more. Crystallographic analysis of 14-3-3 proteins has led to the elucidation of their three-dimensional topology, and the identification of the domains that are involved in their dimerization and in their interaction with ligands.
-
The function of 14-3-3 proteins in the brain remains obscure, but they seem to participate in various physiological cellular processes such as signalling, cell growth, division, adhesion, differentiation, apoptosis and regulation of ion channels.
-
The presence of 14-3-3 proteins in the cerebrospinal fluid of people with Creutzfeldt–Jakob disease has prompted the suggestions that these proteins might be involved in the pathogenesis of this condition and that they might serve as disease biomarkers. Although it is not clear yet whether these suggestions correspond to reality, 14-3-3 proteins have also been implicated in other conditions such as Alzheimer's and Parkinson's diseases, and spinocerebellar ataxia type 1.
-
Future studies should focus on defining the precise roles of 14-3-3 proteins in neuronal physiology, and should try to obtain evidence for a causal relationship between the activity of 14-3-3 proteins and the disease states in which they have been implicated.
Abstract
14-3-3 proteins are abundantly expressed in the brain and have been detected in the cerebrospinal fluid of patients with different neurological disorders. Although the function of this family of highly conserved proteins is not completely known, recent evidence indicates their involvement in multiple cellular processes. By their interaction with more than 100 binding partners, 14-3-3 proteins modulate the action of proteins that are involved in cell cycle and transcriptional control, signal transduction, intracellular trafficking and regulation of ion channels. The study of some of these interactions is sheding light on the role of 14-3-3 proteins in processes such as apoptosis and neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yaffe, M. B. How do 14-3-3 proteins work? — Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57 (2002). A structure-derived model of different forms of 14-3-3-ligand interaction.
van Hemert, M. J., Steensma, H. Y. & van Heusden, G. P. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23, 936–946 (2001).
Tzivion, G., Shen, Y. H. & Zhu, J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20, 6331–6338 (2001).
Moore, B. W. & Perez V. J. in Physiological and Biochemical Aspects of Nervous Integration (ed. Carlson, F. D.) 343–359 (Prentice-Hall, Englewood Cliffs, New Jersey, 1967). First description of 14-3-3 proteins in mammalian brains.
Martin, H. et al. Antibodies against the major brain isoforms of 14-3-3 protein. An antibody specific for the N-acetylated amino-terminus of a protein. FEBS Lett. 331, 296–303 (1993).
Aitken, A., Howell, S., Jones, D., Madrazo, J. & Patel, Y. 14-3-3 α and δ are the phosphorylated forms of raf-activating 14-3-3 β and ζ. In vivo stoichiometric phosphorylation in brain at a Ser-Pro-Glu-Lys motif. J. Biol. Chem. 270, 5706–5709 (1995).
Boston, P. F., Jackson, P. & Thompson, R. J. Human 14-3-3 protein: radioimmunoassay, tissue distribution, and cerebrospinal fluid levels in patients with neurological disorders. J. Neurochem. 38, 1475–1482 (1982).
Pawson, T. & Scott, J. D. Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075–2080 (1997).
Baxter, H. C. et al. Specific 14-3-3 isoform detection and immunolocalization in prion diseases. Biochem. Soc. Trans. 30, 387–391 (2002).
Fu, H., Subramanian, R. R. & Masters, S. C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 (2000). Together, references 1, 2 and 10 provide excellent reviews discussing structure, function and the roles of 14-3-3 proteins in signal transdcution, cell-cycle control and apoptosis.
Liu, D. et al. Crystal structure of the ζ isoform of the 14-3-3 protein. Nature 376, 191–194 (1995). Together with reference 24, this is the first identification of the crystal structure of 14-3-3 isoforms and implications of the inherent functional importance.
Xiao, B. et al. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376, 188–191 (1995).
Petosa, C. et al. 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 273, 16305–16310 (1998).
Wang, H., Zhang, L., Liddington, R. & Fu, H. Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3ζ disrupt its interaction with Raf-1 kinase. J. Biol. Chem. 273, 16297–16304 (1998).
Wang, W. & Shakes, D. C. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43, 384–398 (1996).
Brunet, A. et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmatic transport. J. Cell Biol. 156, 817–828 (2002).
Aitken, A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol. Biol. 50, 993–1010 (2002).
Muslin, A. J., Tanner, J. W., Allen, P. M. & Shaw, A. S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 (1996).
Yaffe, M. B. et al. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91, 961–971 (1997). First identification of the structural basis of specific interactions between 14-3-3s and its target proteins.
Truong, A. B., Masters, S. C., Yang, H. & Fu, H. Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins 49, 321–325 (2002).
Obsil, T., Ghirlando, R., Klein, D. C., Ganguly, S. & Dyda, F. Crystal structure of the 14-3-3ζ: serotonin N-acetyltransferase complex: a role for scaffolding in enzyme regulation. Cell 105, 257–267 (2001).
Ganguly, S. et al. Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc. Natl Acad. Sci. USA 98, 8083–8088 (2001).
Jones, D. H., Ley, S. & Aitken, A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adaptor proteins. FEBS Lett. 368, 55–58 (1995). First evidence that 14-3-3 proteins form homo- and heterodimers with pioneering implications for different modes of action.
Vincenz, C. & Dixit, V. M. 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J. Biol. Chem. 271, 20029–20034 (1996).
Ferl, R. J., Manak, M. S. & Reyes, M. F. The 14-3-3s. Genome Biol. 3, R3010.1–3010.7 (2002).
Rosenquist, M., Sehnke, P., Ferl, R. J., Sommarin, M. & Larsson, C. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51, 446–458 (2000). Detailed examination of 14-3-3 isoforms and species specificity.
Aitken, A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 6, 341–347 (1996).
Niu, J. et al. RGS3 interacts with 14-3-3 via the N-terminal region distinct from the RGS (regulator of G-protein signalling) domain. Biochem. J. 365, 677–684 (2002).
Toska, K. et al. Regulation of tyrosine hydroxylase by stress-activated protein kinases. J. Neurochem. 83, 775–783 (2002).
Zhu, P. et al. The interaction between ADAM 22 and 14-3-3ζ: regulation of cell adhesion and spreading. Biochem. Biophys. Res. Commun. 301, 991–999 (2003).
Bunney, T. D., van den Wijngaard, P. W. & de Boer, A. H. 14-3-3 protein regulation of proton pumps and ion channels. Plant Mol. Biol. 50, 1041–1051 (2002).
Fantl, W. J. et al. Activation of Raf-1 by 14-3-3 proteins. Nature 371, 612–614 (1994).
Irie, K. et al. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science 265, 1716–1719 (1994).
Morrison, D. K. & Cutler, R. E. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9, 174–179 (1997).
Williams, N. G. & Roberts, T. M. Signal transduction pathways involving the Raf proto-oncogene. Cancer Metastasis Rev. 13, 105–116 (1994).
Yip-Schneider, M. T. et al. Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J. 351, 151–159 (2000).
Braselmann, S. & McCormick, F. Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J. 14, 4839–4848 (1995).
Yamamori, B. et al. Purification of a Ras-dependent mitogen-activated protein kinase kinase kinase from bovine brain cytosol and its identification as a complex of B-Raf and 14-3-3 proteins. J. Biol. Chem. 270, 11723–11726 (1995).
Xing, H., Kornfeld, K. & Muslin, A. J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr. Biol. 7, 294–300 (1997).
Kiryu, S., Morita, N., Ohno, K., Maeno, H. & Kiyama, H. Regulation of mRNA expression involved in Ras and PKA signal pathways during rat hypoglossal nerve regeneration. Brain Res. Mol. Brain Res. 29, 147–156 (1995).
Dhillon, A. S. et al. A Raf-1 mutant that dissociates MEK/extracellular signal-regulated kinase activation from malignant transformation and differentiation but not proliferation. Mol. Cell. Biol. 23, 1983–1993 (2003).
Dhaka, A. et al. The RAS effector RIN1 modulates the formation of aversive memories. J. Neurosci. 23, 748–757 (2003).
Namikawa, K., Su, Q., Kiryu-Seo, S. & Kiyama, H. Enhanced expression of 14-3-3 family members in injured motoneurons. Brain Res. Mol. Brain Res. 55, 315–320 (1998).
Cavet, M. E., Lehoux, S. & Berk, B. C. 14-3-3β is a p90 ribosomal S6 kinase isoform 1 (RSK1) binding protein that negatively regulates RSK kinase activity. J. Biol. Chem. 16, 18376–18386 (2003).
Goossens, J. et al. Expression of protein kinase C inhibitor blocks cerebellar long-term depression without affecting Purkinje cell excitability in alert mice. J. Neurosci. 21, 5813–5823 (2001).
Ling, D. S. et al. Protein kinase Mζ is necessary and sufficient for LTP maintenance. Nature Neurosci. 5, 295–296 (2002).
Colombo, P. J., Wetsel, W. C. & Gallagher, M. Spatial memory is related to hippocampal subcellular concentrations of calcium-dependent protein kinase C isoforms in young and aged rats. Proc. Natl Acad. Sci. USA 94, 14195–14199 (1997).
Routtenberg, A., Cantallops, I., Zaffuto, S., Serrano, P. & Namgung, U. Enhanced learning after genetic overexpression of a brain growth protein. Proc. Natl Acad. Sci. USA 97, 7657–7662 (2000).
Dai, J. G. & Murakami K. Constitutively and autonomously active protein kinase C associated with 14-3-3ζ in the rodent brain. J. Neurochem. 84, 23–34 (2003).
Chen, D. -H. et al. Missense mutations in the regulatory domain of PKCγ: a new mechanism for dominant nonepisodic cerebellar ataxia. Am. J. Hum. Genet. 72, 839–949 (2003).
Zha, J., Harada, H., Yang, E., Jockel, J. & Korsmeyer, S. J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL Cell 87, 619–628 (1996).
Nomura, M. et al. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J. Biol. Chem. 278, 2058–2065 (2003).
Zhang, L., Chen, J. & Fu, H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl Acad. Sci. USA 96, 8511–8515 (1999).
Brunet, A. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999).
Xing, H., Zhang, S., Weinheimer, C., Kovacs, A. & Muslin, A. J. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 19, 349–358 (2000).
Basu, S., Totty, N. F., Irwin, M. S., Sudol, M. & Downward, J. Akt phophorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 (2003).
Muslin, A. J. & Xing, H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12, 703–709 (2000). Comprehensive review on the regulation of subcellular localization of ligands by 14-3-3 proteins.
Seimiya, H. et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19, 2652–2661 (2000).
Zhang, P., Chan, S. L., Fu, W., Mendoza, M. & Mattson, M. P. TERT suppresses apoptotis at a premitochondrial step by a mechanism requiring reverse transcriptase activity and 14-3-3 protein binding ability. FASEB J. 17, 767–769 (2003).
Kimura, M. T. et al. 14-3-3 is involved in p75 neurtorphin receptor-mediated signal transduction. J. Biol. Chem. 276, 17291–17300 (2001).
Marra, M. et al. The 30-kilodalton protein present in purified fusicoccin receptor preparations is a 14-3-3-like protein. Plant Physiol. 106, 1497–501 (1994).
Booij, P. P., Roberts, M. R., Vogelzang, S. A., Kraayenhof, R. & De Boer, A. H. 14-3-3 proteins double the number of outward-rectifying K+ channels available for activation in tomato cells Plant J. 20, 673–683 (1999).
Chan, H. C. et al. Modulation of the Ca2+-activated Cl− channel by 14-3-3ε. Biochem. Biophys. Res. Commun. 270, 581–587 (2000).
Zhou, Y. et al. A dynamically regulated 14-3-3, Slob, and Slowpoke potassium channel complex in Drosophila presynaptic nerve terminals. Neuron 22, 809–818 (1999).
Sugita, W., Karoulias, N., Aitken, A. & Ashley, R. H. Chloride intracellular channel protein (CLIC4) (p64H1) binds directly to braindynamin I in a complex containing actin, tubulin and 14-3-3 isoforms. Biochem. J. 359, 55–64 (2001).
Kagan, A., Melman, Y. F., Krumerman, A. & McDonald, T. V. 14-3-3 amplifies and prolongs adrenergic stimulation of HERG K+ channel activity. EMBO J. 21, 1889–1898 (2002).
Rajan, S. et al. Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J. Physiol. (Lond.) 545, 13–26 (2002). An excellent study determining a new mode of action of 14-3-3 proteins in mammalian brains in the control of membrane localization and functional expression of ion channels.
Goldstein, S. A. N. et al. Potassium leak channels and the KCNK family of two-P-domain subunits. Nature Rev. Neurosci. 2, 175–184 (2001).
Lopez, C. J. A new TASK for 14-3-3 proteins. Nature Rev. Neurosci. 3, 915 (2002).
Kim, Y., Bang, H. & Kim, D. TASK-3 a new member of the tandem pore K+ channel family. J. Biol. Chem. 275, 9340–9347 (2000).
Rajan, S. et al. TASK-3, a novel tandem pore domain acid-sensitive K+ channel — an extracellular histidine as pH sensor. J. Biol. Chem. 275, 16650–16657 (2000).
Skoulakis, E. M. & Davis, R. L. 14-3-3 proteins in neuronal development and function. Mol. Neurobiol. 16, 269–284 (1998).
Peyril, A., Weitzdoerfer, R., Gulesserian, T., Fountoulakis, M. & Lubec G. Aberrant expression of signaling-related proteins 14-3-3γ and RACK1 in fetal Down syndrome brain (trisomy 21). Electrophoresis 23, 152–157 (2002).
Kato, M. & Dobyns, W. B. Lissencephaly and the molecular basis of neuronal migration. Hum. Mol. Genet. 12, R89–R96 (2003).
Toyo-oka, K. et al. 14-3-3ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nature Genet. 34, 274–285 (2003).
Will, R. G. Epidemiology of Creutzfeld-Jakob disease. Br. Med. Bull. 49, 960–970 (1993).
Kretzschmar, H. A., Ironside, J. W., DeArmond, S. J. & Tateishi, J. Diagnostic criteria for sporadic Creutzfeldt–Jakob disease. Arch. Neurol. 53, 913–920 (1996).
Harrington, M. G., Merril, C. R., Asher, D. M. & Gajdusek, D. C. Abnormal proteins in the cerebrospinal fluid of patients with Creutzfeldt–Jakob disease. N. Engl. J. Med. 315, 279–283 (1986).
Hsich, G., Kenney, K., Gibbs, C. J., Lee, K. H. & Harrington, M. G. The 14-3-3 brain protein in cerebrospinal fluid as a marker for transmissible spongiform encephalopathies. N. Engl. J. Med. 335, 24–30 (1996). First identification of 14-3-3 proteins as a CSF marker for transmissible spongiform encephalopathies and development of a simple immunoassay for rapid detection.
Zerr, I. et al. Detection of 14-3-3 protein in the cerebrospinal fluid supports the diagnosis of Creutzfeldt–Jakob disease. Ann. Neurol. 43, 32–40 (1998).
Beaudry, P. et al. 14-3-3 protein, neuron-specific enolase, and S-100 protein in cerebrospinal fluid of patients with Creutzfeldt–Jakob disease. Dement. Geriatr. Cogn. Disord. 10, 40–46 (1999).
Brandel, J. P., Delasnerie-Laupretre, N., Laplanche, J. L., Hauw, J. J. & Alperovitch, A. Diagnosis of Creutzfeldt–Jakob disease: effect of clinical criteria on incidence estimates. Neurology 54, 1095–1099 (2000).
Lemstra, A. W. et al. 14-3-3 testing in diagnosing Creutzfeldt–Jakob disease: a prospective study in 112 patients. Neurology 55, 514–516 (2000).
World Health Organization. Global surveillance, diagnosis and therapy of human transmissible spongiform encephalopathies: Report of a WHO consultation WHO/EMC/ZDI/98/9 (World Health Organization, Geneva, 1998). Incorporation of a positive CSF 14-3-3 result into the standard diagnostic criteria for sporadic CJD as a convenient and more sensitive alternative to the typical periodic EEG pattern.
Burkhard, P. R., Sanchez, J. C., Landis, T. & Hochstrasser, D. F. CSF detection of the 14-3-3 protein in unselected patients with dementia. Neurology 56, 1528–1533 (2001).
Will, R. G. et al. Diagnosis of new variant of Creutzfeldt–Jakob disease. Ann. Neurol. 47, 575–582 (2000).
Green, A. J. et al. Use of 14-3-3 and other brain-specific proteins in CSF in the diagnosis of variant Creutzfeldt–Jakob disease. J. Neurol. Neurosurg. Psychiatry 70, 744–748 (2001).
Green, A. J., Ramljak, S., Muller, W. E., Knight, R. S. & Schroder H. C. 14-3-3 in the cerebrospinal fluid of patients with variant and sporadic Creutzfeldt-Jakob disease measured using capture assay able to detect low levels of 14-3-3 protein. Neurosci. Lett. 324, 57–60 (2002).
Collins, S. et al. Creutzfeld–Jakob diease: diagnostic utility of 14-3-3 immunodetection in cerbrospinal fluid. J. Clin. Neurosci. 7, 203–208 (2000).
Haik, S. et al. Dementia with Lewy bodies in a neuropathologic series of suspected Creutzfeldt–Jakob disease. Neurology 55, 1401–1404 (2000).
Kenney, K. et al. An enzyme-linked immunosorbent assay to quantify 14-3-3 proteins in the cerebrospinal fluid of suspected Creutzfeldt–Jakob disease patients. Ann. Neurol. 48, 395–398 (2000).
Wiltfang, J. et al. Isoform pattern of 14-3-3 proteins in the cerebrospinal fluid of patients with Creutzfeldt–Jakob disease. J. Neurochem. 73, 2485–2490 (1999).
Baxter, H. C., Liu, W. G., Forster, J. L., Aitken, A. & Fraser, J. R. Immunolocalisation of 14-3-3 isoforms in normal and scrapie-infected murine brain. Neuroscience 109, 5–14 (2002).
Richard, M. et al. Immunohistochemical localization of 14.3.3ζ protein in amyloid plaques in human spongiform encephalopathies. Acta Neuropathol. 105, 296–302 (2003).
Pietromonaco, S. F., Seluja, G. A., Aitken, A. & Elias, L. Association of 14-3-3 proteins with centrosomes. Blood Cells Mol. Dis. 22, 225–237 (1996).
Martin, H., Rostas, J., Patel, Y. & Aitken, A. Subcellular localisation of 14-3-3 isoforms in rat brain using specific antibodies. J. Neurochem. 63, 2259–2265 (1994).
Jones, D. H. et al. Expression and structural analysis of 14-3-3 proteins. J. Mol. Biol. 245, 375–384 (1995).
Moya, K. L. et al. Immunolocalization of the cellular prion protein in normal brain. Microsc. Res. Tech. 50, 58–65 (2000).
Goedert, M. Tau protein and the neurofibrillary pathology of Alzheimer's disease. Trends Neurosci. 16, 460–465 (1993).
Alonso, A. C., Zaidi, T., Grundke-Iqbal, I. & Iqbal, K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl Acad. Sci. USA 91, 5562–5566 (1994).
Hirokawa, N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 6, 74–81 (1994).
Lee, V. M. Disruption of the cytoskeleton in Alzheimer's disease. Curr. Opin. Neurobiol. 5, 663–668 (1995).
Lee, V. M., Goedert, M. & Trojanowski, J. Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159 (2001).
Wang, J. Z., Gong, C. X., Zaidi, T., Grundke-Iqbal, I. & Iqbal, K. Dephosphorylation of Alzheimer paired helical filaments by protein phosphatase-2A and -2B. J. Biol. Chem. 270, 4854–4860 (1995).
Hashiguchi, M., Sobue, K. & Paudel, H. K. 14-3-3ζ is an effector of tau protein phosphorylation. J. Biol. Chem. 275, 25247–25254 (2000).
Fountoulakis, M., Cairns, N. & Lubec, G. Increased levels of 14-3-3γ and ε proteins in brain of patients with Alzheimer's disease and Down syndrome. J. Neural. Transm. Suppl. 57, 323–335 (1999).
Layfield, R. et al. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci. Lett. 209, 57–60 (1996).
Paudel, H. K. Phosphorylation by neuronal cdc2-like protein kinase promotes dimerization of Tau protein in vitro . J. Biol. Chem. 272, 28328–28334 (1997).
Sun, W. et al. Glycogen synthase kinase-3β is complexed with tau protein in brain microtubules. J. Biol. Chem. 277, 11933–11940 (2002).
Agarwal-Mawal, A. et al. 14-3-3 connects glycogen synthase kinase-3β to tau within a brain microtubule-associated tau phosphorylation complex. J. Biol. Chem. 11, 12722–12728 (2003). In vivo and in vitro study on the possible pathophysiological relevance of 14-3-3 proteins on tau phosphorlylation. 14-3-3ζ was shown to be part of a tau-phosphorylation complex in bovine brain and suggested to facilitate tau phophorylation.
Luo, Z. et al. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383, 181–185 (1996).
Dent, P., Jelinek, T., Morrison, D. K., Weber, M. J. & Sturgill, T. W. Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science 268, 1902–1906 (1995).
Tschampa, H. J. et al. Patients with Alzheimer's disease and dementia with Lewy bodies mistaken for Creutzfeldt–Jakob disease. J. Neurol. Neurosurg. Psychiatry 71, 33–39 (2001).
Hughes, A. J., Ben-Shlomo, Y., Daniel, S. E. & Lees, A. J. What features improve the accuracy of clinical diagnosis in Parkinson's disease: a clinicopathologic study. Neurology 42, 1142–1146 (1992).
Jellinger, K. A. Pathology of Parkinson's disease. Changes other than the nigrostriatal pathway. Mol. Chem. Neuropathol. 14, 153–197 (1991).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging 24, 197–211 (2003).
McKeith. I. G. et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology. 47, 1113–1124 (1996).
Kawamoto, Y. et al. 14-3-3 proteins in Lewy bodies in Parkinson disease and diffuse Lewy body disease brains. J. Neuropathol. Exp. Neurol 61, 245–253 (2002).
Ubl, A. et al. 14-3-3 protein is a component of Lewy bodies in Parkinson's disease–mutation analysis and association studies of 14-3-3η. Brain Res. Mol. Brain Res. 108, 33–39 (2002).
Berg, D., Riess, O. & Bornemann, A. Specification of 14-3–3 proteins in Lewy bodies. Ann. Neurol. 53, 135 (2003).
Ostrerova, N. et al. α-Synuclein shares physical and functional homology with 14-3-3 proteins. J. Neurosci. 19, 5782–5791 (1999).
Ichimura, T. et al. Molecular cloning fo c-DNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc. Natl Acad. Sci. USA 85, 7084–7088 (1988).
Yamauchi, T., Nakata, H. & Fuijsawa, H. A new activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+-, calmodulin-dependent protein kinase. Purification and characterization. J. Biol. Chem. 256, 5404–5409 (1981).
Ichimura, T. et al. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+-, calmodulin-dependent protein kinase II. FEBS Lett. 219, 79–82 (1987). First study to assign a specific role to 14-3-3 proteins.
Kleppe, R., Toska, K. & Haavik, J. Interaction of phosphorylated tyrosine hydroxylase with 14-3-3 proteins: evidence for a phosphoserine 40-dependent association. J. Neurochem. 77, 1097–1107 (2001).
Perez, R. G. et al. A role for α-synuclein in the regulation of dopamine biosynthesis. J. Neurosci. 22, 3090–3099 (2002).
Welch, K. & Yuan, J. Releasing the nerve cell killers. Nature Med. 8, 564–565 (2002).
Xu, J. et al. Dopamine-dependent neurotoxicity of α-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nature Med. 8, 600–606 (2002). Important study indicating a mechanism of selective neurodegeneration of dopaminergic cells of the substantia nigra in PD. α-Synuclein is shown to sequester 14-3-3 protein in this part of the brain, thereby freeing up proteins that promote apoptosis.
Kruger, R. et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nature Genet. 18, 106–108 (1998).
Polymeropoulos, M. H., et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 276, 2045–2047 (1997).
Holzmann, C. et al. Polymorphisms of the α-synuclein promoter: expression analyses and association studies in Parkinson's disease. J. Neural Transm. 110, 67–76 (2003).
Krüger, R. et al. Increased susceptibility to sporadic Parkinson's disease by a certain combined α-synuclein/apolipoprotein E genotype. Ann. Neurol. 45, 611–617 (1999).
Paulson, H. L. et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 (1997).
Skinner, P. J. et al. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389, 971–974 (1997).
Holmberg, M. et al. Spinocerebellar ataxia type 7 (SCA7): a neurodegenerative disorder with neuronal intranuclear inclusions. Hum. Mol. Genet. 7, 913–918 (1998).
Chen, H. -K. et al. Interaction of Akt-phophorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 (2003).
McCampbell, A. et al. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 9, 2197–2202 (2000).
Shimohata, T. et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nature Genet. 26, 29–36 (2000).
Grozinger, C. M. & Schreiber, S. L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3 dependent cellular localization. Proc. Natl Acad. Sci. USA 97, 7835–7840 (2000).
Dunah, A. W. et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296, 2238–2243 (2002).
Steffan, J. S. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila . Nature 413, 739–743 (2001).
Grove, V. E. Jr, Quintanilla, J. & DeVaney, G. T. Improvement of Huntington's disease with olanzapine and valproate. New Engl. J. Med. 343, 973–974 (2000).
Malaspina, A., Kaushik, N. & de Belleroche J. A 14-3-3 mRNA is up-regulated in amyotrophic lateral sclerosis spinal cord. J. Neurochem. 75, 2511–2520 (2000).
Wakabayashi, H. et al. Increased concentrations of 14-3-3 ε, γ and ζ isoforms in cerebrospinal fluid of AIDS patients with neuronal destruction. Clin. Chim. Acta. 312, 97–105 (2001).
Miller, R. F., Green, A. J., Giovannoni, G. & Thompson, E. J. Detection of 14-3-3 brain protein in cerebrospinal fluid of HIV infected patients. Sex Transm. Infect. 76, 408 (2000).
Zerr, I. et al. Current clinical diagnosis in Creutzfeldt–Jakob disease: identification of uncommon variants. Ann. Neurol. 48, 323–329 (2000).
Rosenmann, H. et al. Detection of 14-3-3 protein in the CSF of genetic Creutzfeldt–Jakob disease. Neurology 49, 593–595 (1997).
Satoh, J., Kurohara, K., Yukitake, M. & Kuroda, Y. The 14-3-3 protein detectable in the cerebrospinal fluid of patients with prion-unrelated neurological diseases is expressed constitutively in neurons and glial cells in culture. Eur. Neurol. 41, 216–225 (1999).
Saiz, A., Marin, C., Tolosa, E. & Graus, F. Diagnostic usefulness of the determination of protein 14-3-3 in cerebrospinal fluid in Creutzfeldt–Jakob disease. Neurologia 13, 324–328 (1998).
Will, R. G. et al. Cerbrospinal fluid test for new-variant Creutzfeldt–Jakob disease. Lancet 348, 955 (1996).
Saiz, A. et al. Detection of 14-3-3 brain protein in the cerebrospinal fluid of patients with paraneoplastic neurological disorders. Ann. Neurol. 46, 774–777 (1999).
Hernandez Echebarria, L. E. et al. Detection of 14-3-3 protein in the CSF of a patient with Hashimoto's encephalopathy. Neurology 54, 1539–1540 (2000).
Irani, D. N. & Kerr, D. A. 14-3-3 protein in the cerebrospinal fluid of patients with acute transverse myelitis. Lancet 355, 901 (2000).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Kimura, M. Evolutionary rate at the molecular level. Nature 217, 624–626 (1968).
Page, R. D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 (1996).
Perriere, G. & Gouy, M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369 (1996).
Acknowledgements
We thank R. Liddington for his helpful support and for providing figure 1. Our studies on 14-3-3 proteins have been supported by the Fortuene programme of the University of Tübingen and the FORUN programme of the University of Rostock.
Author information
Authors and Affiliations
Corresponding author
Glossary
- DEAE-CELLULOSE CHROMATOGRAPHY
-
Type of ion-exchange chromatography in which the diethylaminoethyl group acts as a weak base, allowing the binding of negatively charged molecules.
- STARCH-GEL ELECTROPHORESIS
-
Form of electrophoresis in which starch is used as solid matrix. It is particularly useful for the separation of the allelic variants of a protein.
- NONSENSE MUTATION
-
A mutation that results in the introduction of a stop codon to cause the premature termination of the protein.
- MISSENSE MUTATION
-
A mutation that results in the substitution of an amino acid in a protein.
- ZINC FINGER
-
A protein module in which cysteine or cysteine–histidine residues coordinate a zinc ion. Zinc fingers are often used in DNA recognition and in protein–protein interactions.
- OUTWARDLY-RECTIFYING K+ CHANNELS
-
Potassium channels through which ions flow more easily out of than into the cell. They are important for membrane repolarization after an action potential.
- DYNAMIN
-
A GTPase that takes part in endocytosis. It seems to be involved in severing the connection between the nascent vesicle and the donor membrane.
- LEAK CHANNELS
-
Proteins that are responsible for the background membrane currents that are present at rest and that rise instantly to a new steady level in response to voltage changes.
- LISSENCEPHALY
-
Literally meaning 'smooth brain', lissencephaly is a human brain disorder that is characterized by absence or reduction of the cerebral convolutions.
- RETT SYNDROME
-
An inherited, X-linked neurological disorder that is lethal to males. In females, the disorder is characterized by rapid neurological deterioration that leads to dementia, autism, loss of speech and voluntary movement, and microcephaly. Rett syndrome is related to mutations in Mecp2, which encodes a methyl-CpG binding protein that acts as a transcriptional repressor.
- RASMUSSEN'S ENCEPHALITIS
-
A progressive neurological disorder characterized by frequent and severe seizures, loss of motor skills and speech, paralysis on one side of the body, inflammation of the brain, dementia and mental deterioration. The disorder, which affects a single cerebral hemisphere, generally occurs in children under the age of 10, and is related to the production of glutamate receptor autoantibodies.
- SCHILDER'S DISEASE
-
Also known as diffuse sclerosis, Schilder's disease is a progressive demyelinating disorder that commonly begins in childhood. Its symptoms include dementia, aphasia, seizures, personality changes, loss of balance, weakness, and vision and speech impairment. This disorder is a variant of multiple sclerosis.
- NEUROLEPTIC
-
This term was originally coined to refer to the effects of early antipsychotic agents on cognition and behaviour.
Rights and permissions
About this article
Cite this article
Berg, D., Holzmann, C. & Riess, O. 14-3-3 proteins in the nervous system. Nat Rev Neurosci 4, 752–762 (2003). https://doi.org/10.1038/nrn1197
Issue Date:
DOI: https://doi.org/10.1038/nrn1197
This article is cited by
-
Transcriptome analysis of the cerebral cortex of acrylamide-exposed wild-type and IL-1β-knockout mice
Archives of Toxicology (2024)
-
14-3-3 proteins regulate cullin 7-mediated Eag1 degradation
Cell & Bioscience (2023)
-
14-3-3γ haploinsufficiency leads to altered dopamine pathway and Parkinson’s disease-like motor incoordination in mice
Molecular Brain (2023)
-
Clinical significance of serum levels of 14-3-3β protein in patients with stable chronic obstructive pulmonary disease
Scientific Reports (2023)
-
14-3-3ζ Mediates GABAAR Activation by Interacting with BIG1
Molecular Neurobiology (2023)